Mission
The Center for Research Computing supports leading-edge research with free access to advanced computing hardware and software for fields across the entire research community, along with training and consultation by CRC research faculty. CRC offers the following services:
- Access to cutting-edge computer hardware and software for enabling transformative research
- Workshops teaching the most effective ways to use Pitt CRC's computing resources
- Personalized consultation on refining projects at the computational code or workflow level

Resources:
- Website: https://crc.pitt.edu
- Getting Started: https://crc.pitt.edu/getting-started
- User Support: https://crc.pitt.edu/UserSupport
Here is a schematic of all the key parts of the advanced computing infrastructure. The users' computer is the Client. h2p.crc.pitt.edu is the remote login server. CRC compute and storage resources are behind a firewall within PittNet.
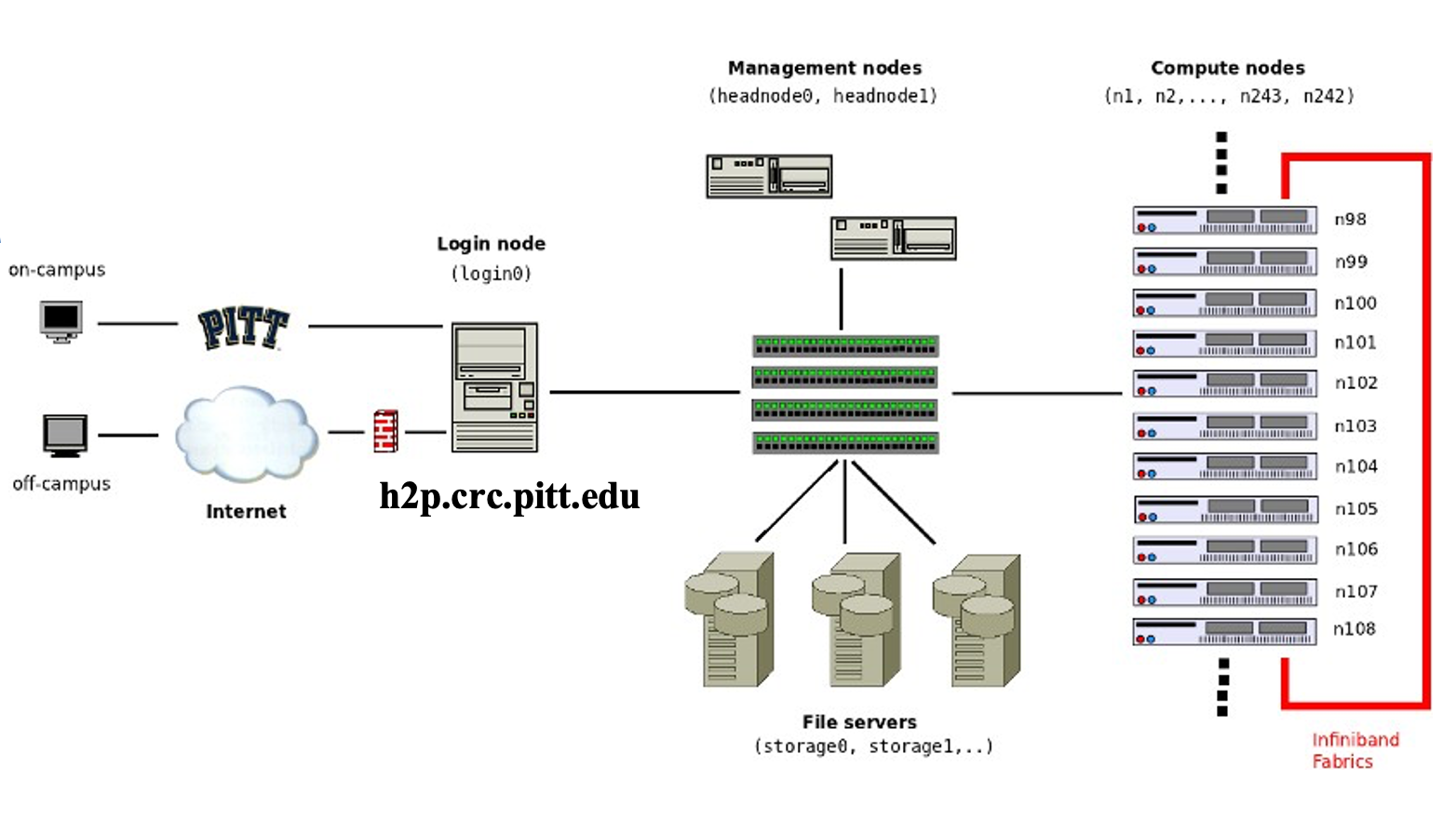
The CRC computing and storage resources reside at the Pitt data center and are firewalled within PittNet. What this means is that you will need to establish a VPN in order to gain access. Pitt offers two VPN tools: (1) Pulse VPN and (2) Global Protect. Both software can be downloaded from software.pitt.edu.
References
We outline the steps and settings each VPN client below.
1. Pulse VPN
Download and run the Pulse installer.
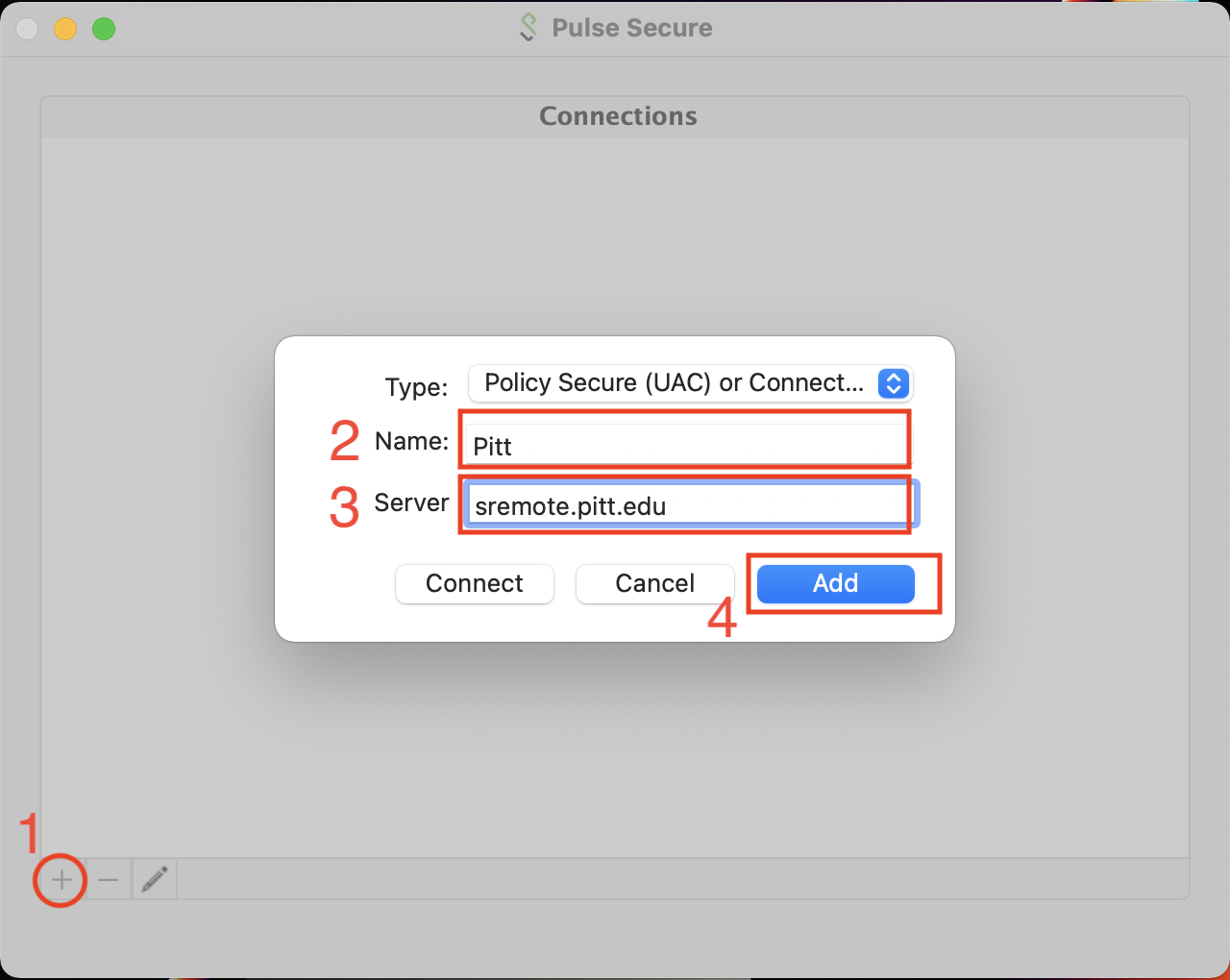
When you first run Pulse Secure, there will be no Connections entries. We want to create a new Connection entry with the Server field set to sremote.pitt.edu. The Name field can be arbitrary but Pitt will work. The type should be set to Policy Secure (UAC).
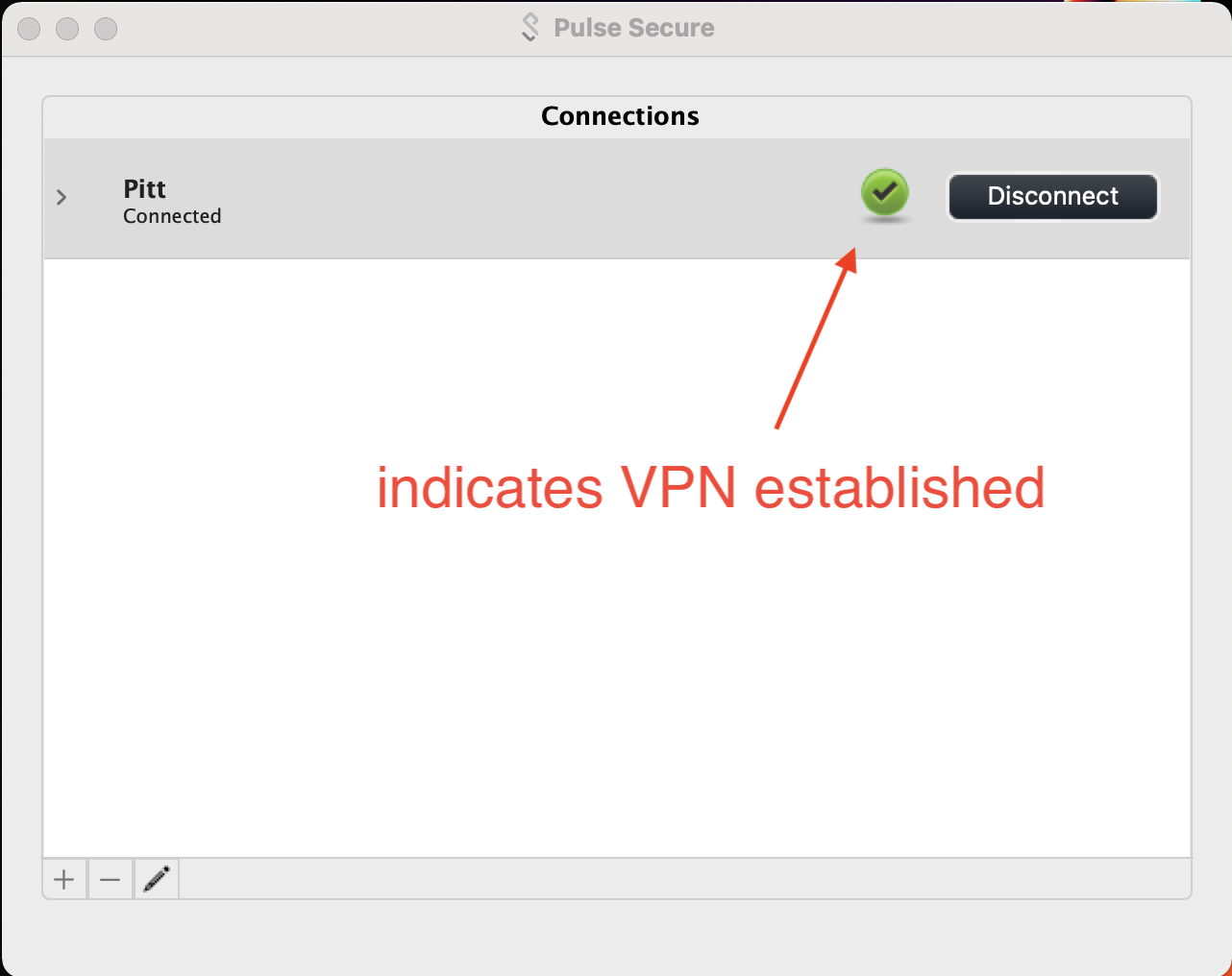
Once you have the Connections entry added, click on the Connect button to initiate the VPN 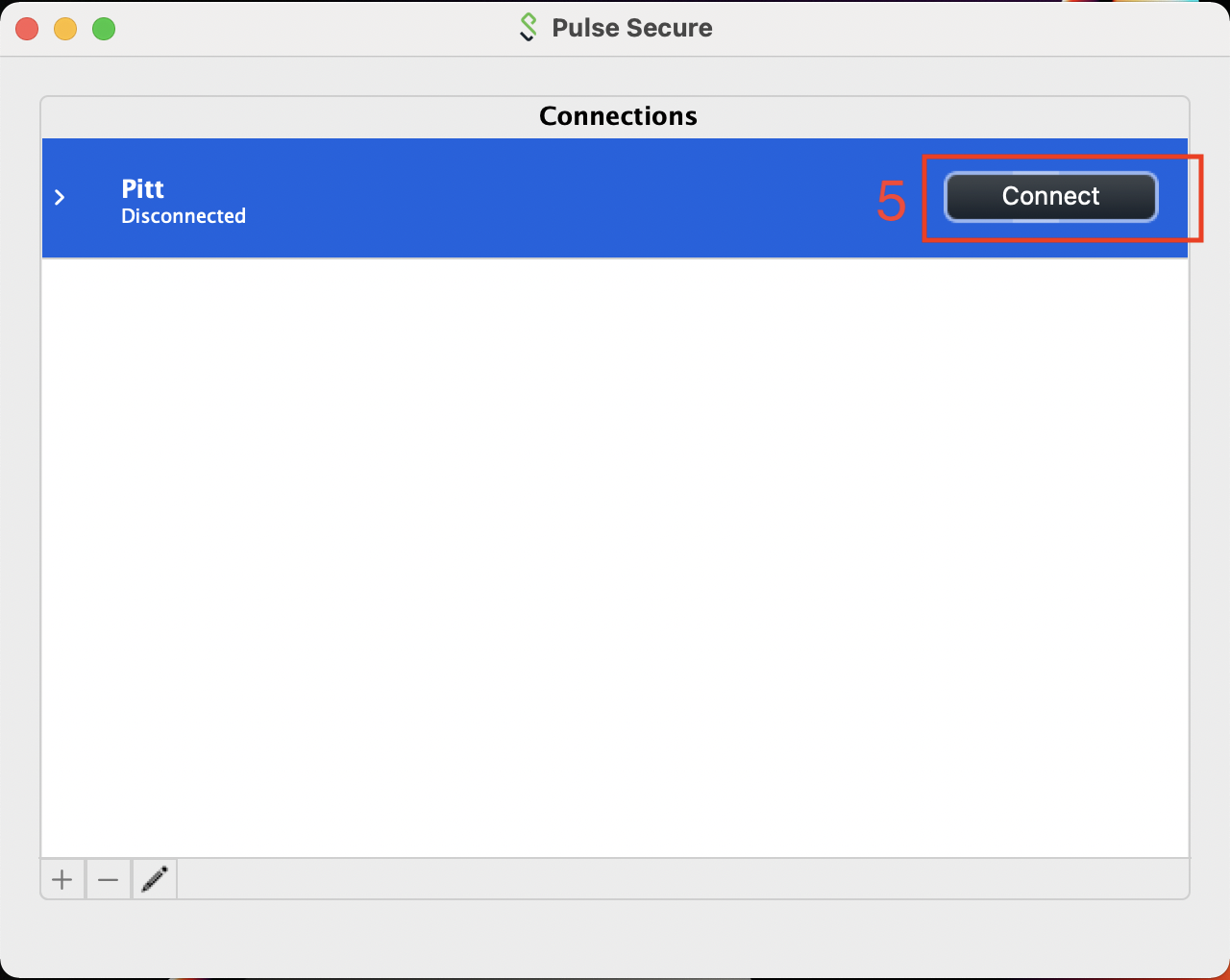
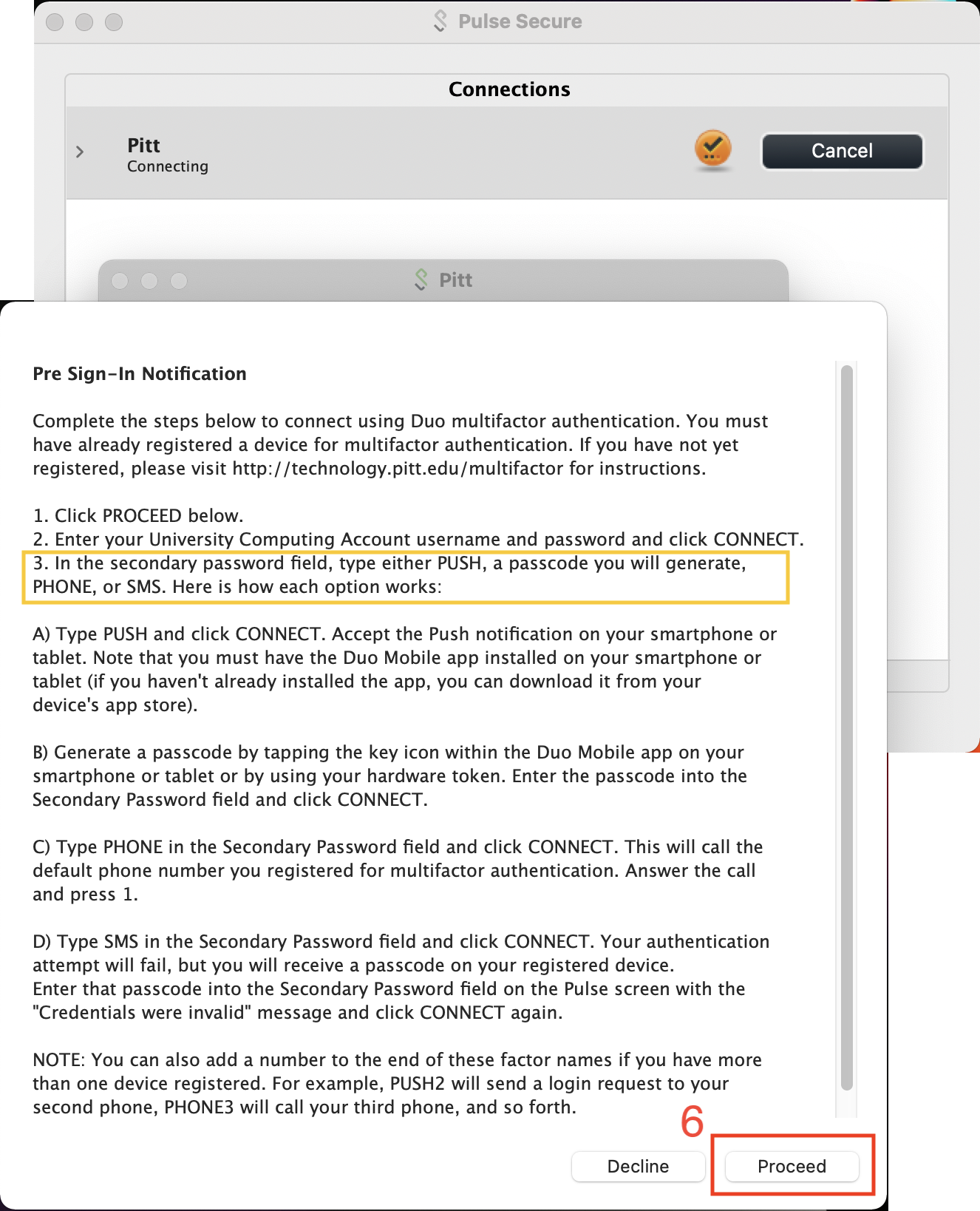
Connect using your Pitt User Name and Password
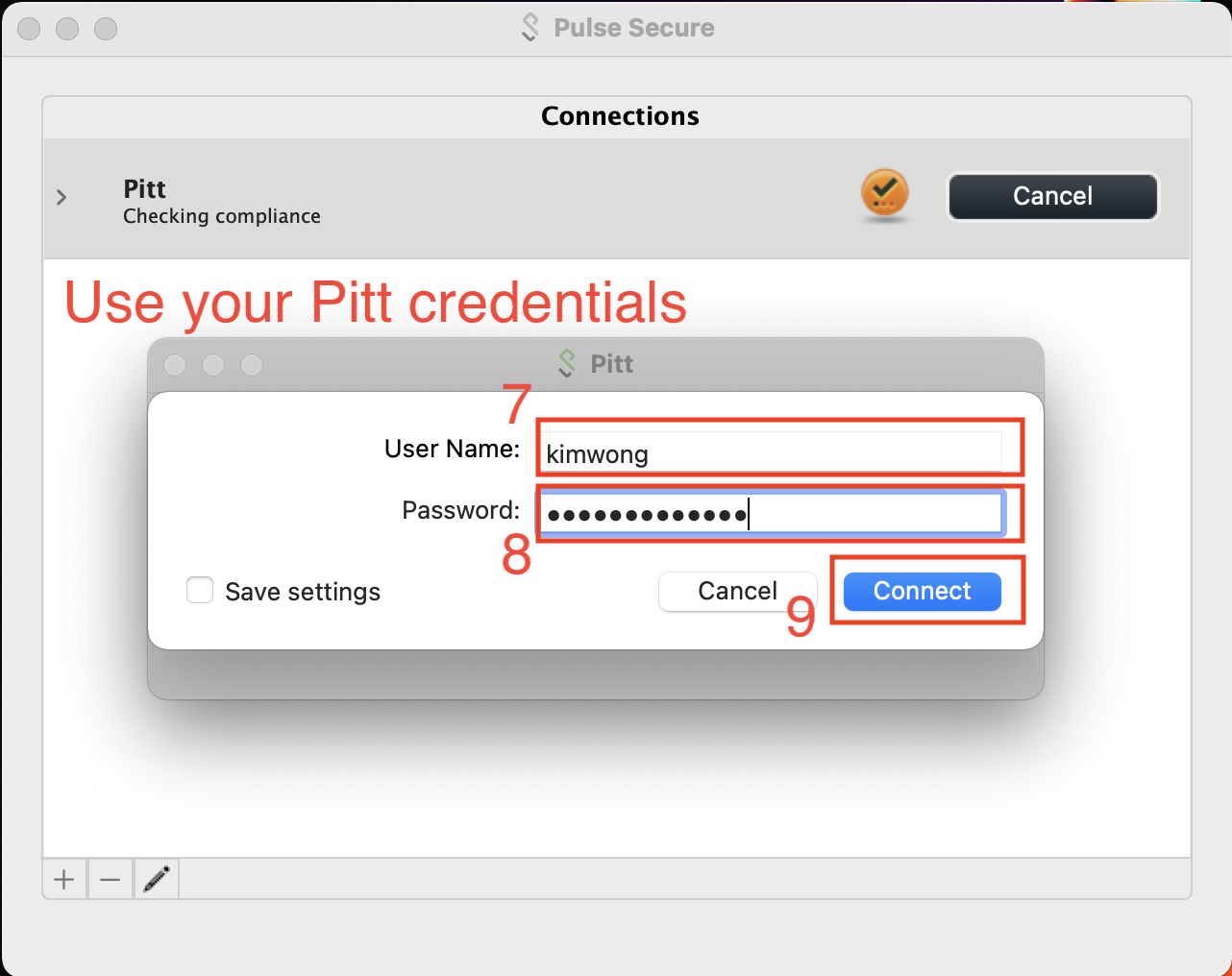
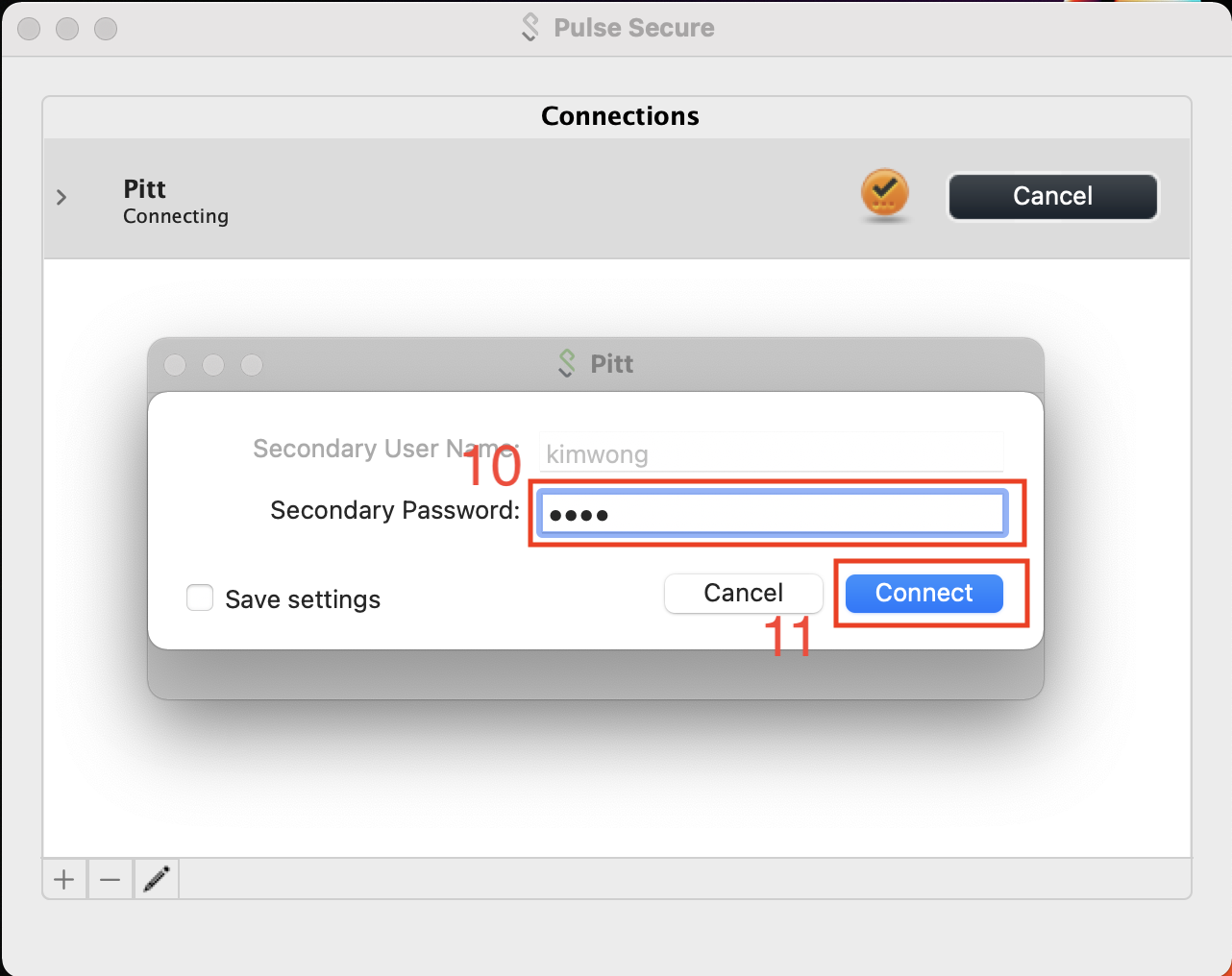
Most Pitt members have set up Duo on their cell phone as the secondary authentication factor. In this case, the Secondary Passworkd is PUSH.
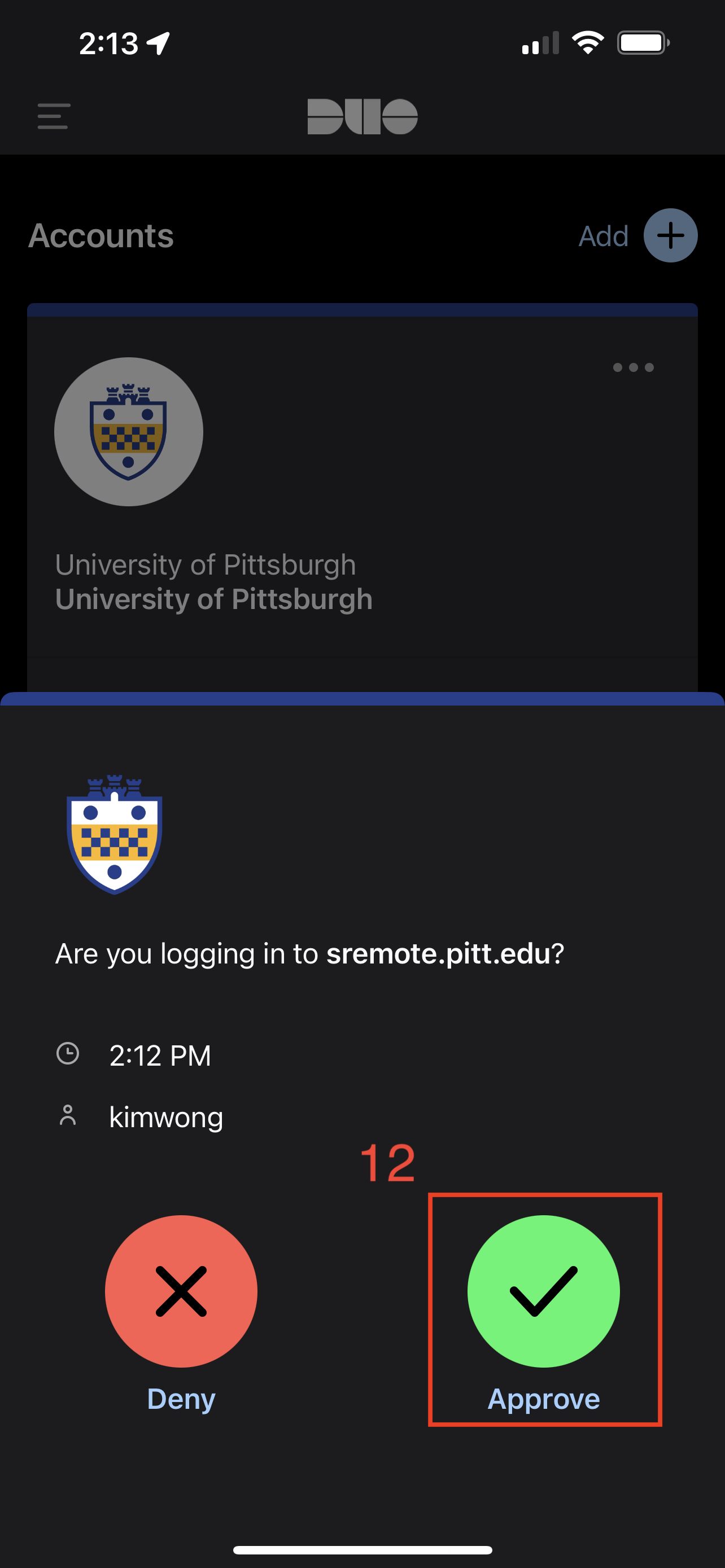
This will prompt for login approval via the Duo app on your phone.
The CRC access role is Firewall-SAM-USERS-Pulse.
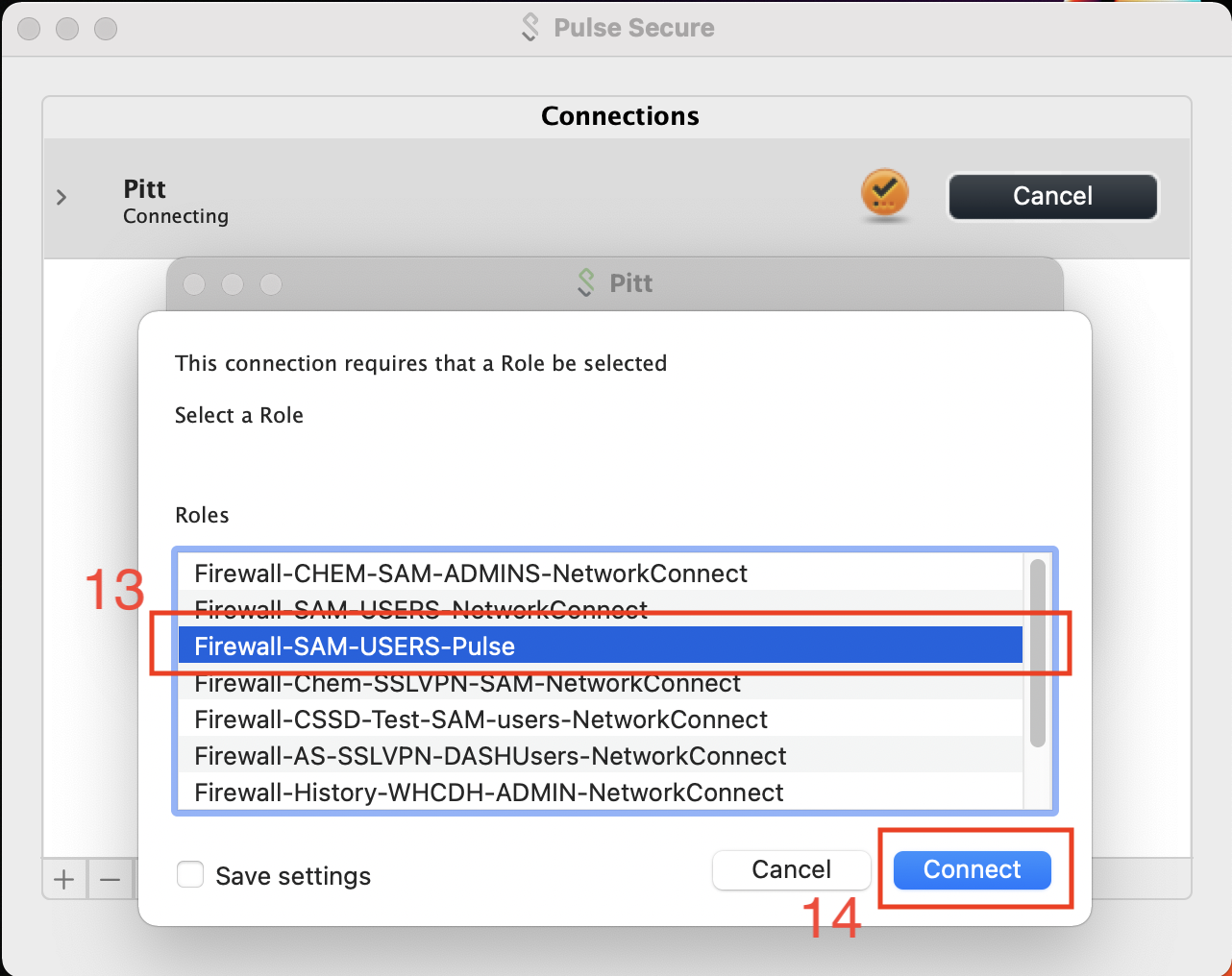
A check within a green sphere indicates successful VPN connection.
2. Global Protect
CRC provides several modes for accessing the advanced computing and storage resoures, including:
- traditional terminal
- Web GUI portal
- Open OnDemand
- JupyterHub
We briefly describe each interface below.
1. The traditional terminal
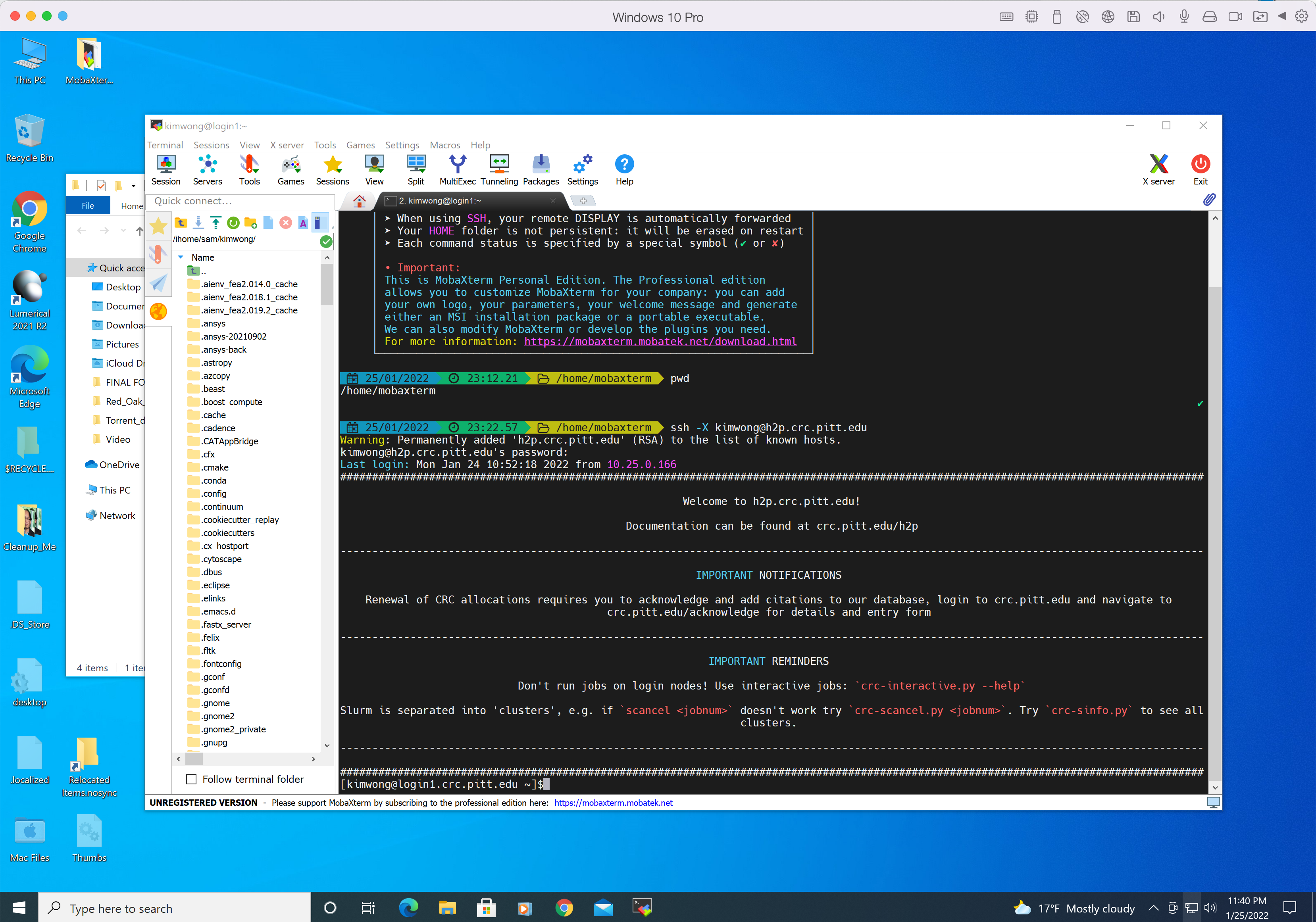
If your client computer is Windows, I recommend downloading the portable edition of MobaXterm. Execute MobaXterm and click on the + Start local terminal button to open an terminal. Recall that in The Ecosystem schematic, the remote login server is h2p.crc.pitt.edu We are going to use ssh to connect to the H2P login node. Here are the connection details:
- connection protocol: ssh
- remote hostname: h2p.crc.pitt.edu
- authentication credentials: Pitt username and password
The syntax to connect to the H2P login node is
ssh -X <username>@h2p.crc.pitt.edu
where <username> is your Pitt username in lowercase and the answer to the prompt is the corresponding password. The -X option enables X forwarding for applications that generate a GUI such as xclock. If you type xclock on the commandline, you should get a clock app showing in Windows. Below is my login session from MobaXterm.
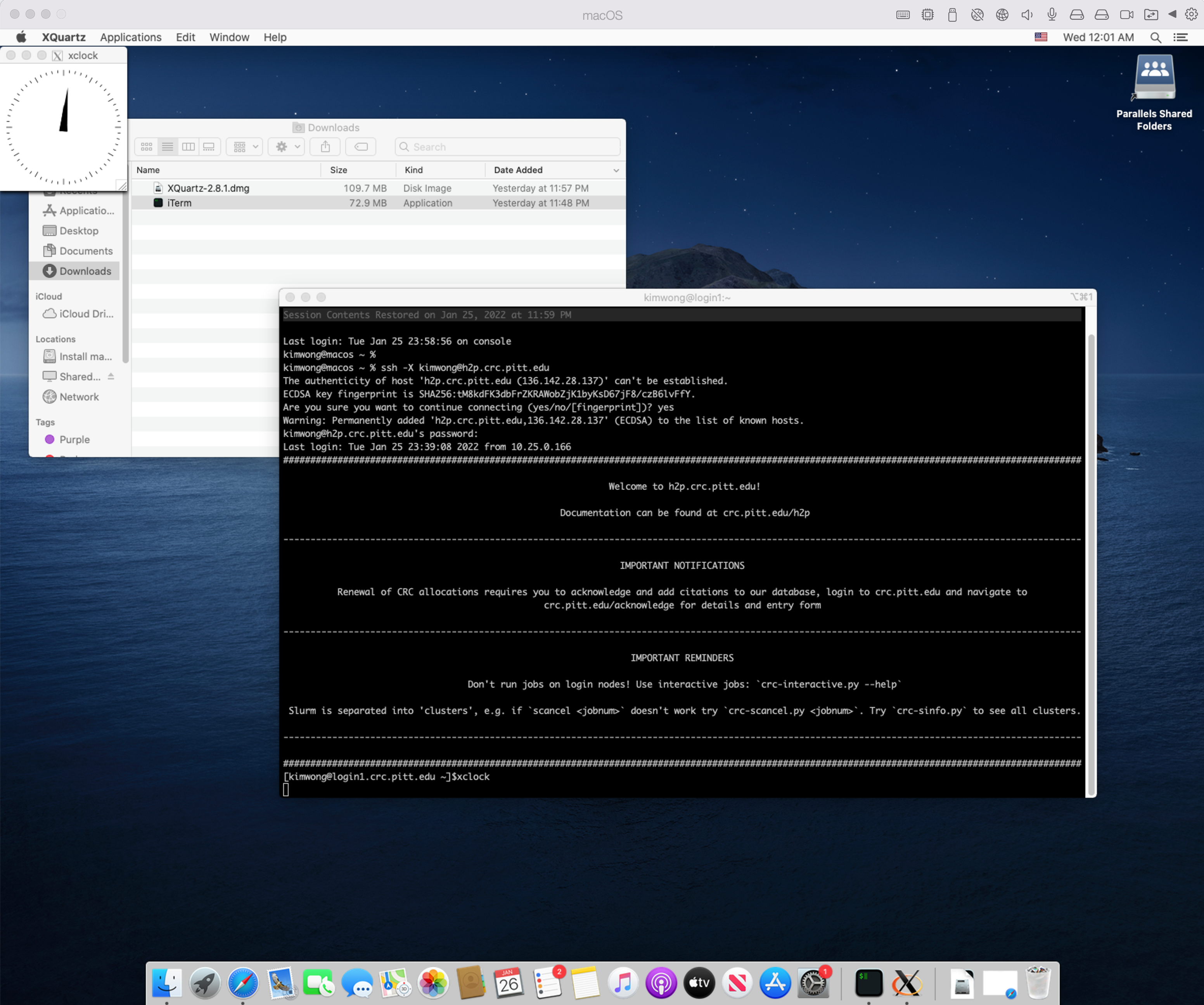
If your client computer is MacOS, I recommend downloading iTerm2. While MacOS already has a builtin Terminal in the Utilities folder, iTerm2 is more feature-rich. iTerm2 is just a terminal. To render graphics, you will need to install XQuartz, which provides the X Server component. Below is my login session using iTerm2 and XQuartz, following the same syntax as shown earlier.
2. Web GUI Portal
CRC provides access to a Linux Desktop using a web browser. Point your browser to viz.crc and authenticate using your Pitt credentials.
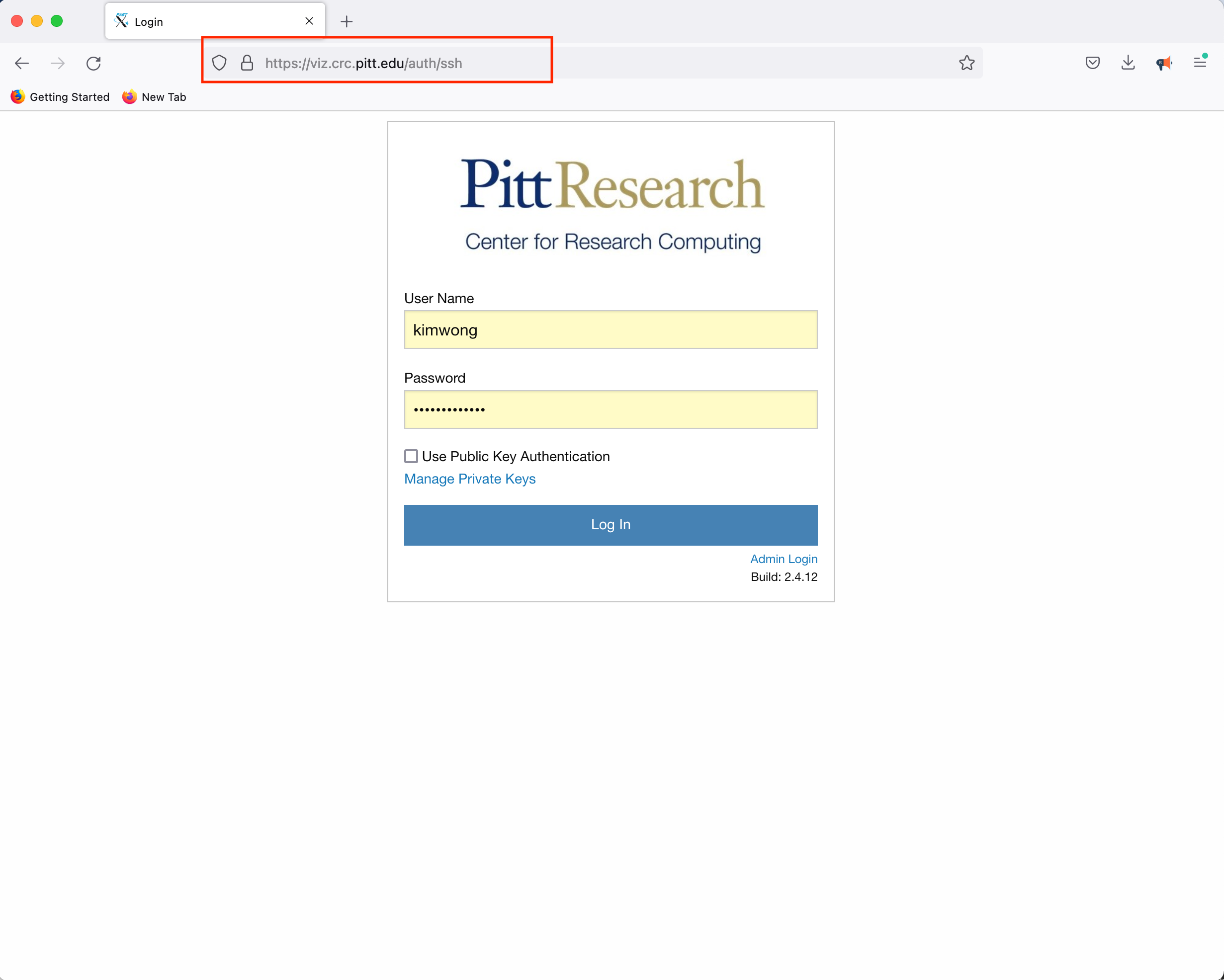
Click Launch Session, click on MATE, and click Launch
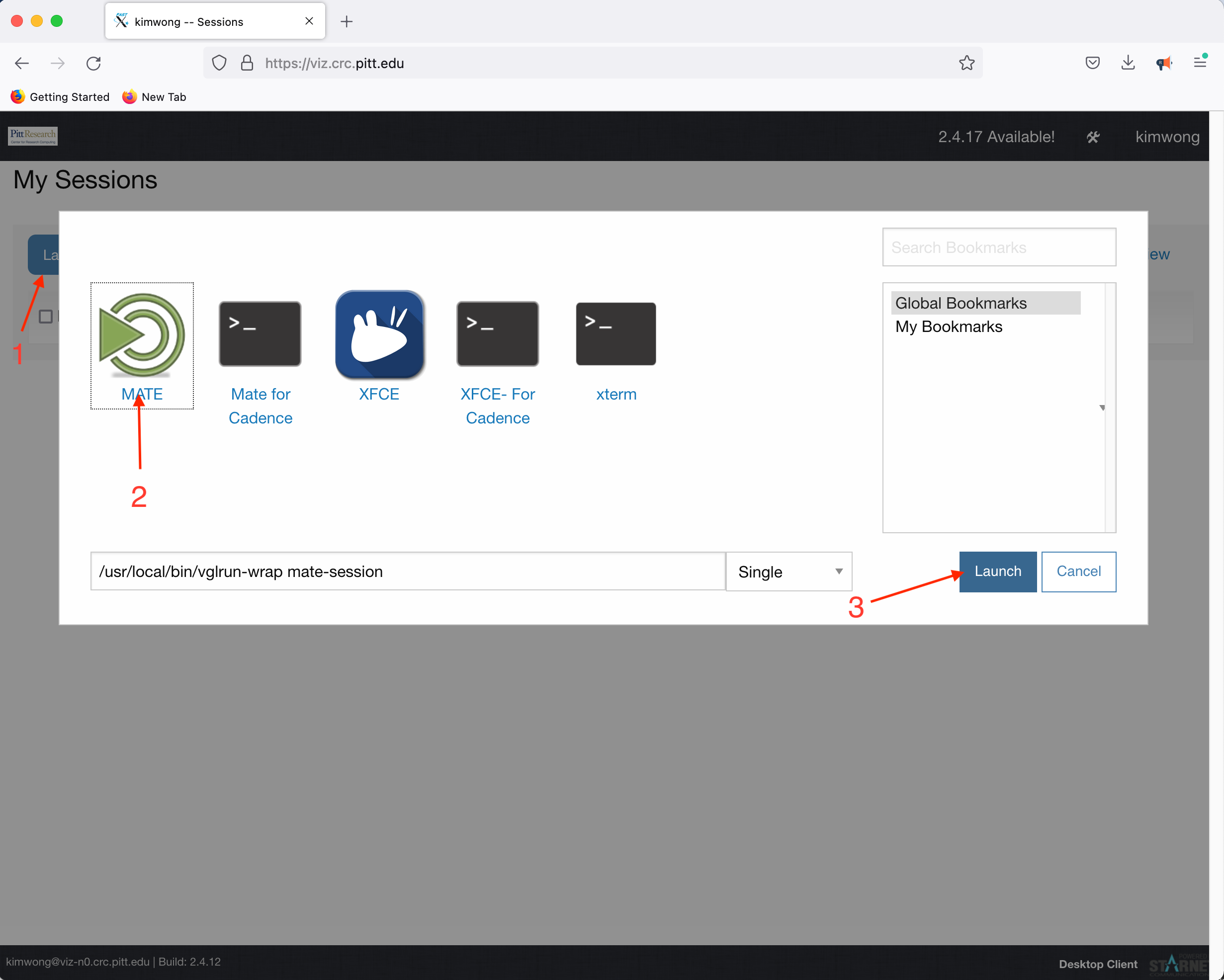
What is presented to you will be a Linux Desktop, with graphical capabilities, where you can interact with the rest of the CRC compute and storage resources
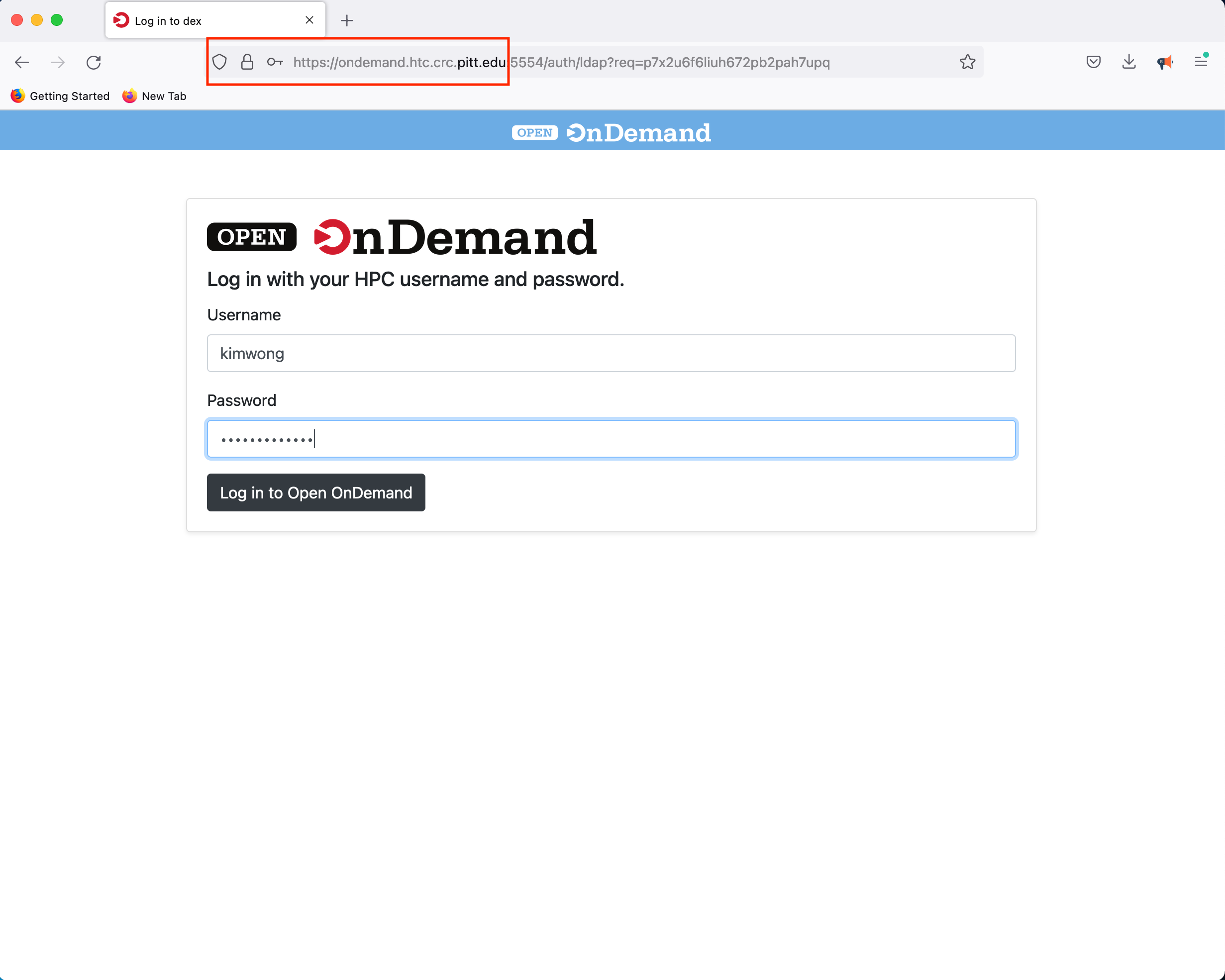
3. Open OnDemand
Similar to viz.crc, our Open OnDemand web portal provides our users access to interactive compute resources. The full documentation for CRC's implementation features are described here. Point your browser to OnDemand and authenticate using your Pitt credentials
Once you log in, you will be presented with a menu of options. For example, click on the Interactive Apps dropdown menu
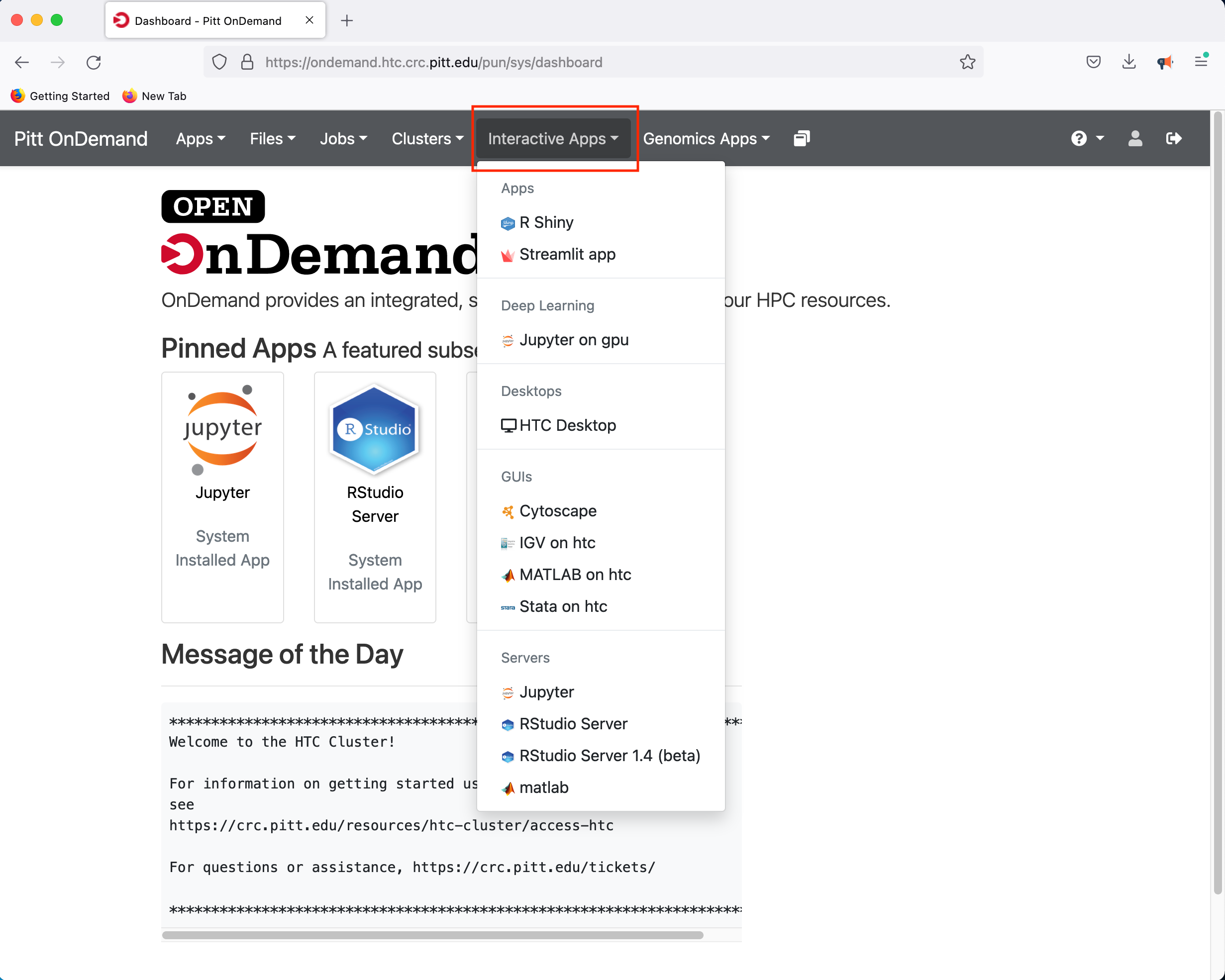
If you select the R Studio Server option, you will be presented with a panel where you can configure the resources to suit your needs
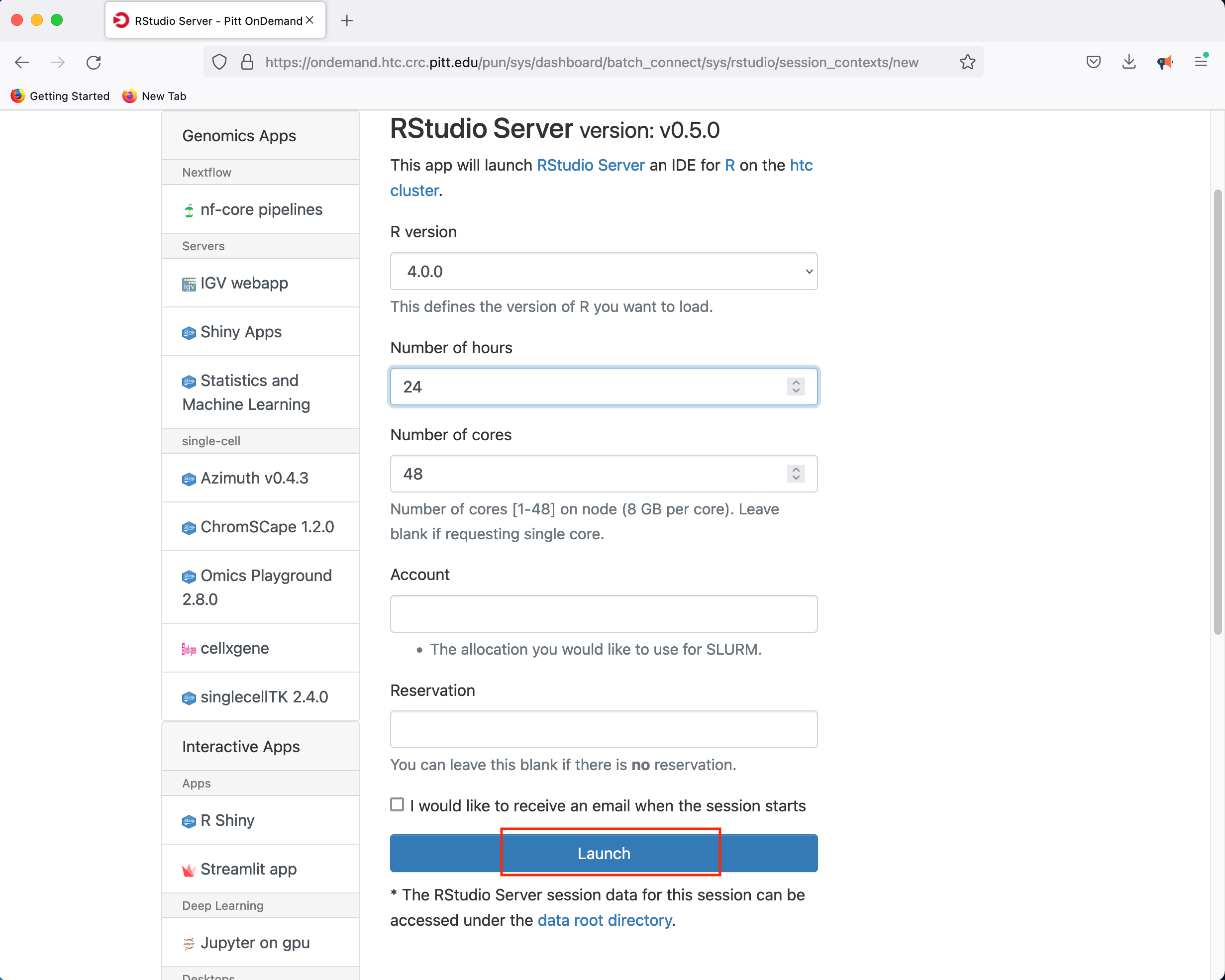
Clicking Launch will submit the resource request to the queue and will present a button to Connect to RStudio Server when the resources have been allocated.
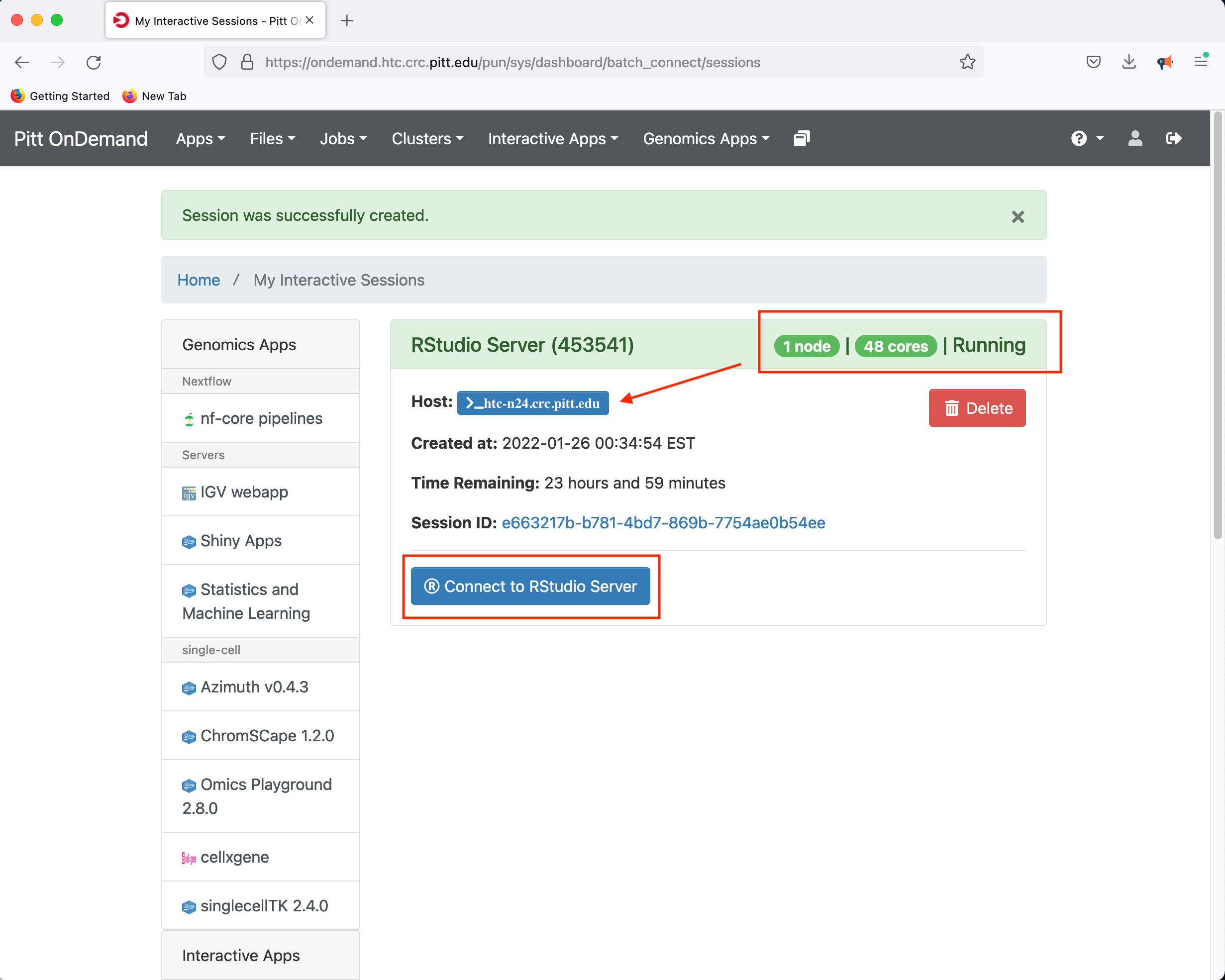
In this instance, the compute node allocated to host the RStudio Server is htc-n24 with 48 cores for a period of 24 hours.
4. JupyterHub
CRC provides a JupyterHub instance in support of teaching. Point your browser to hub.crc and authenticate using your Pitt credentials when presented with the Pitt Passport page. Clicking on Start My Server provides a panel for requesting access to CPUs and GPUs
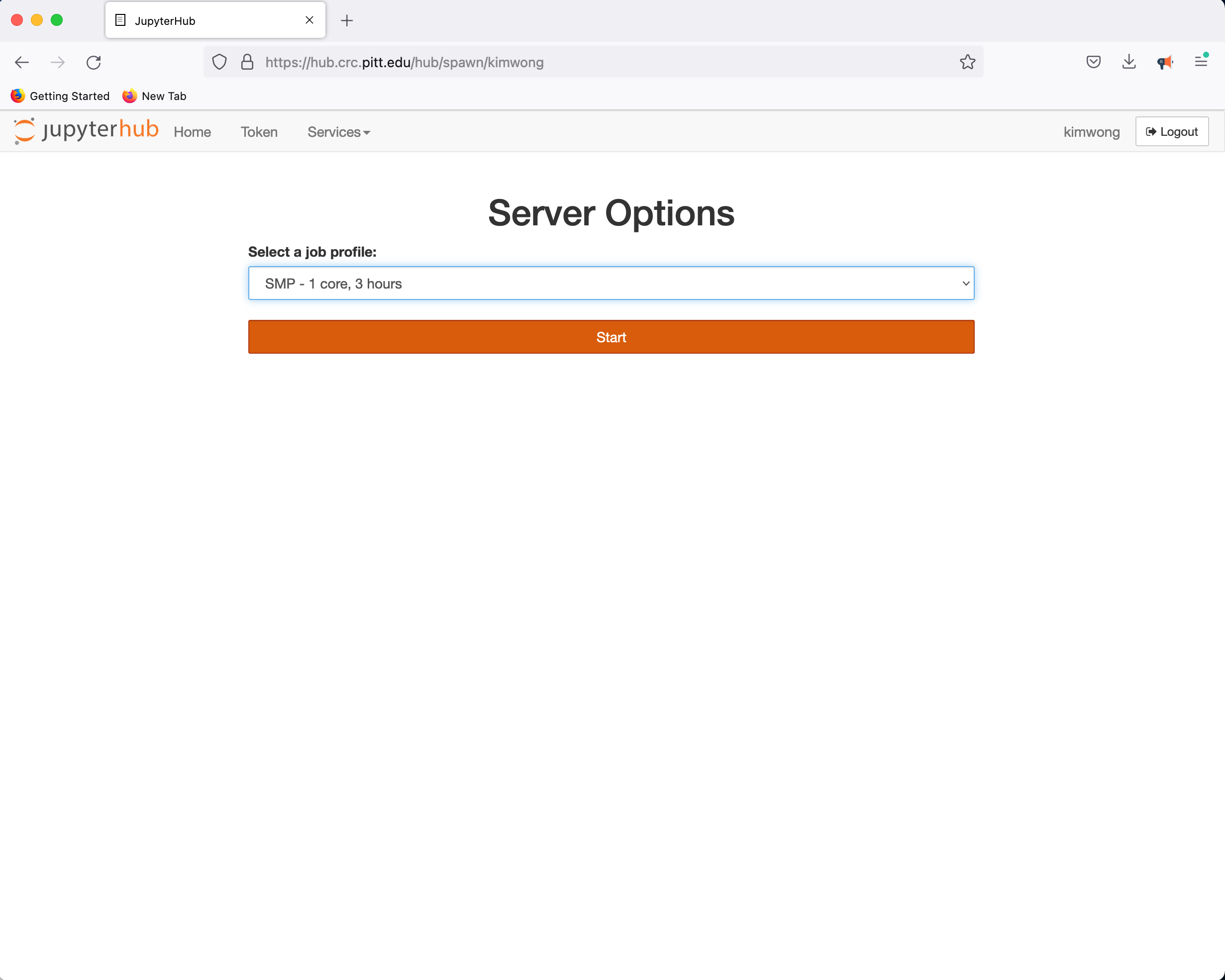
followed by the familiar Python environment
Once you become familiar with the Linux commmandline, the traditional terminal interface will become the most efficient method for accessing the CRC compute and storage resources. I will now log in to the cluster using my testing gnowmik account, which is kimwong spelled backwards and does not have the superuser privileges in my default account
Cheese-Grater:~ kimwong$ ssh -X gnowmik@h2p.crc.pitt.edu
gnowmik@h2p.crc.pitt.edu's password:
Warning: untrusted X11 forwarding setup failed: xauth key data not generated
Last login: Thu Jan 13 12:09:21 2022
#########################################################################################################################################################################################
Welcome to h2p.crc.pitt.edu!
Documentation can be found at crc.pitt.edu/h2p
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
IMPORTANT NOTIFICATIONS
Renewal of CRC allocations requires you to acknowledge and add citations to our database, login to crc.pitt.edu and navigate to crc.pitt.edu/acknowledge for details and entry form
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
IMPORTANT REMINDERS
Don't run jobs on login nodes! Use interactive jobs: `crc-interactive.py --help`
Slurm is separated into 'clusters', e.g. if `scancel <jobnum>` doesn't work try `crc-scancel.py <jobnum>`. Try `crc-sinfo.py` to see all clusters.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
#########################################################################################################################################################################################
[gnowmik@login1 ~]$ pwd
/ihome/kwong/gnowmik
[gnowmik@login1 ~]$ ls
CRC Desktop zzz_cleanmeup
[gnowmik@login1 ~]$
CRC uses the Lmod Environment Modules tool to manage and provision software applications. The command module spider shows if a package is available. For example
[gnowmik@login1 ~]$ module spider python
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
python:
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Description:
Anaconda is the leading open data science platform powered by Python.
Versions:
python/anaconda2.7-4.2.0_westpa
python/anaconda2.7-4.2.0
python/anaconda2.7-4.4.0_genomics
python/anaconda2.7-5.2.0_westpa
python/anaconda2.7-5.2.0
python/anaconda2.7-2018.12_westpa
python/anaconda3.5-4.2.0-dev
python/anaconda3.5-4.2.0
python/anaconda3.6-5.2.0_deeplabcut
python/anaconda3.6-5.2.0_leap
python/anaconda3.6-5.2.0
python/anaconda3.7-5.3.1_genomics
python/anaconda3.7-2018.12_westpa
python/anaconda3.7-2019.03_astro
python/anaconda3.7-2019.03_deformetrica
python/anaconda3.7-2019.03
python/anaconda3.8-2020.11
python/anaconda3.9-2021.11
python/bioconda-2.7-5.2.0
python/bioconda-3.6-5.2.0
python/bioconda-3.7-2019.03
python/intel-3.5
python/intel-3.6_2018.3.039
python/intel-3.6_2019.2.066
python/intel-3.6
python/ondemand-jupyter-python3.8
python/3.7.0-dev
python/3.7.0-fastx
python/3.7.0
Other possible modules matches:
biopython openslide-python
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
To find other possible module matches do:
module -r spider '.*python.*'
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
For detailed information about a specific "python" module (including how to load the modules) use the module's full name.
For example:
$ module spider python/ondemand-jupyter-python3.8
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
[gnowmik@login1 ~]$
shows that we have several versions of Python available. Packages typically have dependencies. To discover these dependencies, apply the module spider command to a specific installed package
[gnowmik@login1 ~]$ module spider python/anaconda3.7-2019.03_deformetrica
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
python: python/anaconda3.7-2019.03_deformetrica
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Description:
Anaconda is the leading open data science platform powered by Python. Compatible with gcc/8.2.0
Other possible modules matches:
biopython, openslide-python
You will need to load all module(s) on any one of the lines below before the "python/anaconda3.7-2019.03_deformetrica" module is available to load.
gcc/8.2.0
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
To find other possible module matches do:
module -r spider '.*python/anaconda3.7-2019.03_deformetrica.*'
[gnowmik@login1 ~]$
If you attempt to directly load the this Python to your session environment, you will encounter an error because the gcc/8.2.0 dependency has not been satisfied
[gnowmik@login1 ~]$ module load python/anaconda3.7-2019.03_deformetrica
Lmod has detected the following error: These module(s) exist but cannot be loaded as requested: "python/anaconda3.7-2019.03_deformetrica"
Try: "module spider python/anaconda3.7-2019.03_deformetrica" to see how to load the module(s).
[gnowmik@login1 ~]$
Load the gcc/8.2.0 module first before loading the desired Python
[gnowmik@login1 ~]$ module load gcc/8.2.0
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) gcc/8.2.0
[gnowmik@login1 ~]$ module load python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$
You can also load all the packages using a single commandline, making sure that the dependencies are to the left of the package
[gnowmik@login1 ~]$ module purge
[gnowmik@login1 ~]$ module load gcc/8.2.0 python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$
Now let's backup and fill in our knowledge gap regarding two commands that were introduced under the radar. They are module purge and module list. These commands do exactly as the words imply. module list is to list all the loaded software packages and module purge is to get rid of all the packages from your session environment
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module purge
[gnowmik@login1 ~]$ module list
No modules loaded
[gnowmik@login1 ~]$
Now, you might be wondering if it is possible to remove a particular package while keeping others. Why don't we try it
[gnowmik@login1 ~]$ module purge
[gnowmik@login1 ~]$ module list
No modules loaded
[gnowmik@login1 ~]$ module spider matlab
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
matlab:
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Description:
Matlab R2021b
Versions:
matlab/R2015a
matlab/R2017a
matlab/R2018a
matlab/R2019b
matlab/R2020b
matlab/R2021a
matlab/R2021b
Other possible modules matches:
matlab-mcr matlab-proxy
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
To find other possible module matches do:
module -r spider '.*matlab.*'
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
For detailed information about a specific "matlab" module (including how to load the modules) use the module's full name.
For example:
$ module spider matlab/R2021b
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
[gnowmik@login1 ~]$ module load matlab/R2021b
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) fontconfig/2.10.95 2) matlab/R2021b
[gnowmik@login1 ~]$ module load gcc/8.2.0 python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) fontconfig/2.10.95 2) matlab/R2021b 3) openmpi/3.1.1 4) gcc/8.2.0 5) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module rm matlab/R2021b
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$
In the above commands, I loaded Matlab R2021b and then Python. Notice that Matlab can be loaded directly but that there is also a side effect of automatically loading fontconfig/2.10.95. Next, I loaded the gcc/8.2.0 dependecy before the specific Python package. This Python package also has a side effect of automatically loading openmpi/3.1.1. Lastly, when I unload the Matlab package, matlab/R2021b and fontconfig/2.10.95 are removed from the environment.
You might wonder, What happens if I unload Python instead of Matlab? Let's try it out
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module load matlab/R2021b
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) openmpi/3.1.1 2) gcc/8.2.0 3) python/anaconda3.7-2019.03_deformetrica 4) fontconfig/2.10.95 5) matlab/R2021b
[gnowmik@login1 ~]$ module rm python/anaconda3.7-2019.03_deformetrica
[gnowmik@login1 ~]$ module list
Currently Loaded Modules:
1) fontconfig/2.10.95 2) matlab/R2021b
[gnowmik@login1 ~]$
The effect of unloading a package module is to remove all depedencies and to return the session environment to the state before loading the package. The command to unload a package is module rm . You might also wonder if module unload might be a better choice of words for the command. As a matter of fact, module rm and module unload are synonymous. Try it out.
These are the few commands you need to memorize for manipulating the software package environments. The reason why it is necessary to employ Lmod is because our research user community use a broad range of software applications and not all applications are compatible with each other.
Because CRC operates a shared resource for the Pitt research community, we need a tool to ensure fair and equitable access. CRC uses the SLURM workload manager to manage job submissions. This is a batch queueing system that will allocate resources based on defined policies. What this means is that users will be submitting job scripts to the queue and jobs will run when the SLURM scheduler allocates the requested resources in accordance to scheduling policies. Let's jump right into the details.
Shown below is the architecture of a SLURM job submission script
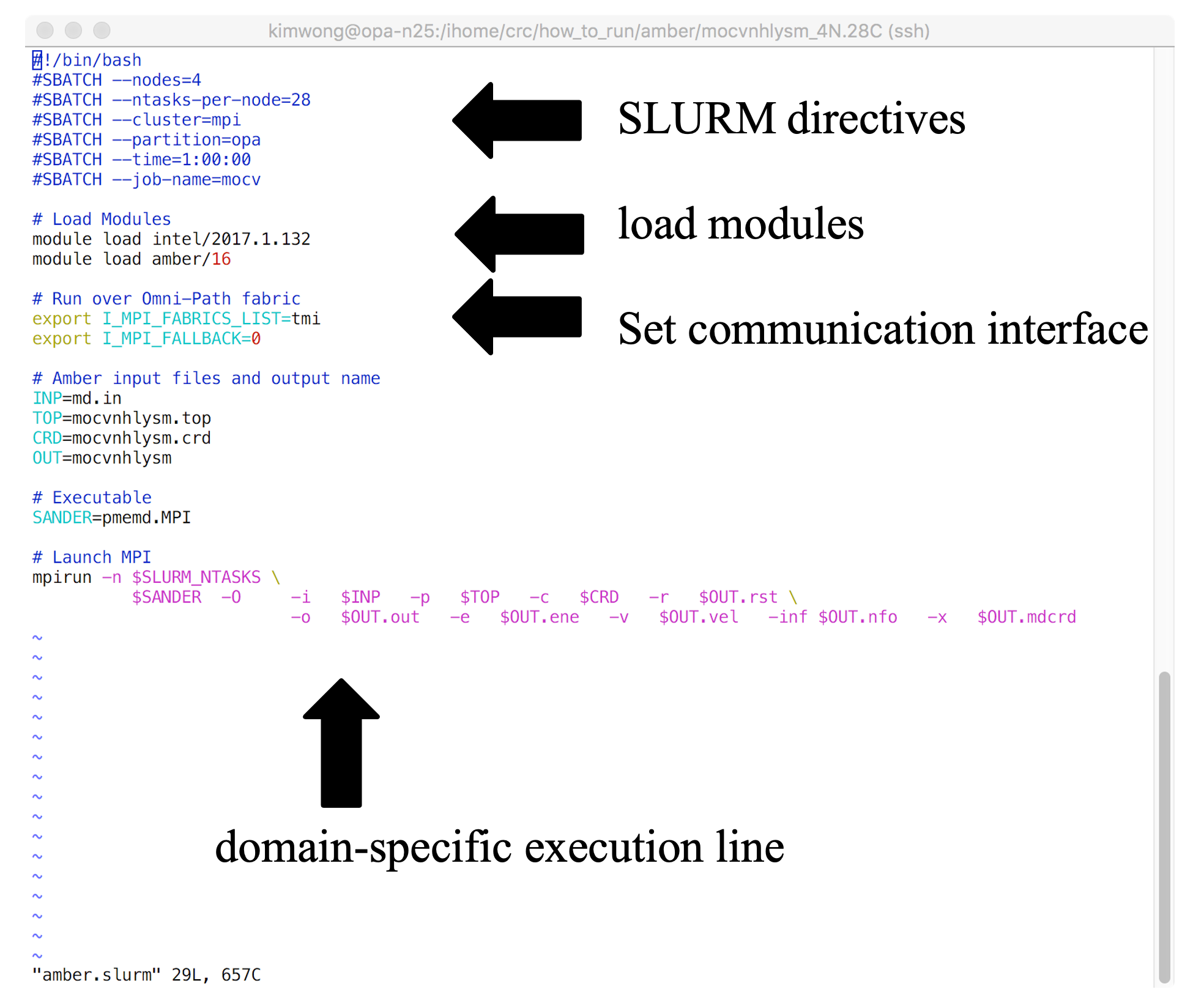
The SLURM job submission script is essentially a text file that contains (1) commands to SLURM, (2) commands to Lmod, (3) any environment settings for communication or software, and (4) the application-specific execution command. The commands execute sequentially line-by-line from top to bottom (unless you background the command with an & at the end). CRC provides a growing number of example job submission scripts for specific software applications
[gnowmik@login1 ~]$ ls /ihome/crc/how_to_run/
abaqus ansys comsol DeepLabCut-1.02 febio gaussian hello-world julia lumerical matlab mopac nektar++ pbdr quantumespresso stata vasp
abm bioeng2370_2021f cp2k deformetrica fluent gpaw hfss lammps lumerical.test molecularGSM mosek openfoam psi4 r tinker westpa
amber blender damask fdtd gamess gromacs ipc lightgbm lumerical.test2 molpro namd orca qchem sas turbomole xilinx
[gnowmik@login1 ~]$
Let's copy a few examples and go from there
[gnowmik@login1 ~]$ cd
[gnowmik@login1 ~]$ pwd
/ihome/kwong/gnowmik
[gnowmik@login1 ~]$ cp -rp /ihome/crc/how_to_run/amber/mocvnhlysm_1N.24C_OMPI_SMP .
[gnowmik@login1 ~]$ cp -rp /ihome/crc/how_to_run/amber/mocvnhlysm_1titanX.1C .
[gnowmik@login1 ~]$ cp -rp /ihome/crc/how_to_run/amber/mocvnhlysm_2GTX1080.2C .
[gnowmik@login1 ~]$ ls
CRC Desktop mocvnhlysm_1N.24C_OMPI_SMP mocvnhlysm_1titanX.1C mocvnhlysm_2GTX1080.2C zzz_cleanmeup
[gnowmik@login1 ~]$
First let's go into the mocvnhlysm_1N.24C_OMPI_SMP directory and show the contents of the SLURM submission script
[gnowmik@login1 ~]$ cd mocvnhlysm_1N.24C_OMPI_SMP
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ ls
amber.slurm logfile md.in mocvnhlysm.crd mocvnhlysm.nfo mocvnhlysm.rst mocvnhlysm.top
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ cat amber.slurm
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=12
#SBATCH --cluster=smp
#SBATCH --partition=high-mem
#SBATCH --time=1:00:00
#SBATCH --job-name=mocv
# Load Modules
module purge
module load gcc/5.4.0
module load openmpi/3.0.0
module load amber/16_gcc-5.4.0
# Run over Omni-Path fabric
#export I_MPI_FABRICS_LIST=tmi
#export I_MPI_FALLBACK=0
# Amber input files and output name
INP=md.in
TOP=mocvnhlysm.top
CRD=mocvnhlysm.crd
OUT=mocvnhlysm
# Executable
SANDER=pmemd.MPI
# Launch MPI
mpirun -n $SLURM_NTASKS \
$SANDER -O -i $INP -p $TOP -c $CRD -r $OUT.rst \
-o $OUT.out -e $OUT.ene -v $OUT.vel -inf $OUT.nfo -x $OUT.mdcrd
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$
The SLURM directives begin with the #SBATCH prefix and instructs the scheduler to allocate 1 node with 12 cores within the high-mem partition on the smp cluster for 1 hour. Then the submission script loads the Amber molecular dynamics package and dependencies, followed by application-specific execution syntax. Use sbatch to submit the job
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ sbatch amber.slurm
Submitted batch job 5103575 on cluster smp
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ squeue -M smp -u $USER
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
5103575 high-mem mocv gnowmik R 0:18 1 smp-512-n1
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ tail mocvnhlysm.out
|---------------------------------------------------
NSTEP = 500 TIME(PS) = 2021.000 TEMP(K) = 300.08 PRESS = 0.0
Etot = -292450.7926 EKtot = 68100.1600 EPtot = -360550.9527
BOND = 534.0932 ANGLE = 1306.5392 DIHED = 1661.1194
1-4 NB = 555.1360 1-4 EEL = 4509.5203 VDWAALS = 51060.9002
EELEC = -420178.2610 EHBOND = 0.0000 RESTRAINT = 0.0000
Ewald error estimate: 0.1946E-03
------------------------------------------------------------------------------
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$
Every job submission is assigned a job id. In this case it is 5103575. Use the squeue command to check on the status of submitted jobs. The -M option is to specify the cluster and the -u flag is used to only output information for a particular username
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ echo $USER
gnowmik
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ squeue -M smp -u $USER
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ tail -30 mocvnhlysm.out
| Total 14.01 5.51
| PME Load Balancing CPU Time, Average for All Tasks:
|
| Routine Sec %
| ------------------------------------
| Atom Reassign 0.01 0.00
| Image Reassign 0.01 0.00
| FFT Reassign 0.01 0.00
| ------------------------------------
| Total 0.02 0.01
| Final Performance Info:
| -----------------------------------------------------
| Average timings for last 0 steps:
| Elapsed(s) = 0.07 Per Step(ms) = Infinity
| ns/day = 0.00 seconds/ns = Infinity
|
| Average timings for all steps:
| Elapsed(s) = 254.36 Per Step(ms) = 50.87
| ns/day = 3.40 seconds/ns = 25436.13
| -----------------------------------------------------
| Master Setup CPU time: 0.54 seconds
| Master NonSetup CPU time: 254.10 seconds
| Master Total CPU time: 254.64 seconds 0.07 hours
| Master Setup wall time: 3 seconds
| Master NonSetup wall time: 254 seconds
| Master Total wall time: 257 seconds 0.07 hours
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$
In the time needed to write the descriptions, the job had completed. If you leave out the -u option to squeue, you get reporting of everyone's jobs on the specified cluster
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ squeue -M smp
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
5046724 smp desf_y_1 sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046730 smp isof_y_1 sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046732 smp enfl_y_1 sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046760 smp enfl_pf_ sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046761 smp enfl_pcl sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046762 smp isof_pcl sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046763 smp isof_poc sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046773 smp desf_pf_ sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046780 smp desf_poc sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046853 smp desf_bo_ sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
5046869 smp isof_bo_ sadowsky PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4304639 smp run_mrs. taa80 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158825 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158826 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158827 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158828 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158829 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158830 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158831 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158832 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158833 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158834 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158835 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158836 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158837 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158838 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158839 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158840 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158841 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158842 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158843 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
3158844 smp methane/ sum57 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4684270 smp reverse has197 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4684271 smp generate has197 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120436 high-mem chr7 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120437 high-mem chr6 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120438 high-mem chr5 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120439 high-mem chr4 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120440 high-mem chr3 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120441 high-mem chr2 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4120443 high-mem chr1 kowaae22 PD 0:00 1 (AssocGrpCPURunMinutesLimit)
4684277 smp reverse has197 PD 0:00 1 (Dependency)
4684278 smp generate has197 PD 0:00 1 (Dependency)
5097014 high-mem eom jmb503 PD 0:00 1 (MaxCpuPerAccount)
4917460_468 smp canP13 ryanp PD 0:00 1 (launch failed requeued held)
5085232 high-mem T2T_CENP mam835 R 2-11:54:39 1 smp-256-n2
5085230 high-mem T2T_CENP mam835 R 2-11:54:49 1 smp-256-n1
5091263 smp bowtie_c sat143 R 9:48:55 1 smp-n192
5080187 high-mem LCuH_dim yuz171 R 1-16:03:36 1 smp-3072-n1
5086871 smp 24-1_17- jsh89 R 1-13:40:04 1 smp-n86
5095388 smp sampled_ sem156 R 1:04:09 1 smp-n20
5095387 smp sampled_ sem156 R 1:23:19 1 smp-n21
5095386 smp sampled_ sem156 R 1:47:10 1 smp-n16
5095385 smp sampled_ sem156 R 2:20:17 1 smp-n5
5095384 smp sampled_ sem156 R 2:23:30 1 smp-n11
5095382 smp sampled_ sem156 R 2:31:08 1 smp-n6
5095378 smp sampled_ sem156 R 3:14:25 1 smp-n3
5089347_250 smp RFshim ans372 R 2:30:41 1 smp-n195
5089347_249 smp RFshim ans372 R 2:31:14 1 smp-n98
5089347_248 smp RFshim ans372 R 2:32:59 1 smp-n152
5089347_247 smp RFshim ans372 R 2:34:46 1 smp-n111
5089347_246 smp RFshim ans372 R 2:35:51 1 smp-n51
Now let's take a look at a job submission script to the gpu cluster
[gnowmik@login1 ~]$ cd
[gnowmik@login1 ~]$ cd mocvnhlysm_1titanX.1C
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ pwd
/ihome/kwong/gnowmik/mocvnhlysm_1titanX.1C
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ ls
amber.slurm md.in mocvnhlysm.crd mocvnhlysm.nfo mocvnhlysm.rst mocvnhlysm.top
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ cat amber.slurm
#!/bin/bash
#SBATCH --job-name=gpus-1
#SBATCH --output=gpus-1.out
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cluster=gpu
#SBATCH --partition=titanx
#SBATCH --gres=gpu:1
#SBATCH --time=24:00:00
# Load Modules
module purge
module load cuda/7.5.18
module load amber/16-titanx
# Amber input files and output name
INP=md.in
TOP=mocvnhlysm.top
CRD=mocvnhlysm.crd
OUT=mocvnhlysm
# Executable
SANDER=pmemd.cuda
# Launch PMEMD.CUDA
echo AMBERHOME $AMBERHOME
echo SLURM_NTASKS $SLURM_NTASKS
nvidia-smi
$SANDER -O -i $INP -p $TOP -c $CRD -r $OUT.rst \
-o $OUT.out -e $OUT.ene -v $OUT.vel -inf $OUT.nfo -x $OUT.mdcrd
[gnowmik@login1 mocvnhlysm_1titanX.1C]$
The content of this job submission script is similar to the one for the smp cluster, with key differences in the SLURM directives and the specification of the GPU-accelerated Amber package and executable. Here, we are requesting 1 node with 1 core and 1 GPU within the titanx partition in the gpu cluster for 24 hours. We submit the job using the sbatch command.
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ sbatch amber.slurm
Submitted batch job 260052 on cluster gpu
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ squeue -M gpu -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260052 titanx gpus-1 gnowmik R 0:06 1 gpu-stage06
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ tail mocvnhlysm.out
------------------------------------------------------------------------------
NSTEP = 1000 TIME(PS) = 2022.000 TEMP(K) = 301.12 PRESS = 0.0
Etot = -292271.3092 EKtot = 68336.6875 EPtot = -360607.9967
BOND = 490.8433 ANGLE = 1305.8711 DIHED = 1690.9079
1-4 NB = 555.5940 1-4 EEL = 4530.8677 VDWAALS = 51423.4399
EELEC = -420605.5206 EHBOND = 0.0000 RESTRAINT = 0.0000
------------------------------------------------------------------------------
[gnowmik@login1 mocvnhlysm_1titanX.1C]$
While this job is running, let's run the other GPU-accelerated example
[gnowmik@login1 mocvnhlysm_1titanX.1C]$ cd ../mocvnhlysm_2GTX1080.2C/
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ cat amber.slurm
#!/bin/bash
#SBATCH --job-name=gpus-2
#SBATCH --output=gpus-2.out
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=2
#SBATCH --cluster=gpu
#SBATCH --partition=gtx1080
#SBATCH --gres=gpu:2
#SBATCH --time=24:00:00
# Load Modules
module purge
module load cuda/8.0.44
module load amber/16-gtx1080
# Amber input files and output name
INP=md.in
TOP=mocvnhlysm.top
CRD=mocvnhlysm.crd
OUT=mocvnhlysm
# Executable
SANDER=pmemd.cuda.MPI
# Launch PMEMD.CUDA
echo AMBERHOME $AMBERHOME
echo SLURM_NTASKS $SLURM_NTASKS
nvidia-smi
mpirun -n $SLURM_NTASKS \
$SANDER -O -i $INP -p $TOP -c $CRD -r $OUT.rst \
-o $OUT.out -e $OUT.ene -v $OUT.vel -inf $OUT.nfo -x $OUT.mdcrd
In this example, we are requesting 2 GPUs and 2 cores on a node within the gtx1080 partition of the gpu cluster. Let's submit the job using sbatch and check on the queue
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ squeue -M gpu -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260052 titanx gpus-1 gnowmik R 6:15 1 gpu-stage06
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ sbatch amber.slurm
Submitted batch job 260053 on cluster gpu
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ squeue -M gpu -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260053 gtx1080 gpus-2 gnowmik R 0:04 1 gpu-n25
260052 titanx gpus-1 gnowmik R 6:23 1 gpu-stage06
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$
You see that we now have two jobs running on the GPU cluster, one on the titanx partition and the other on the gtx1080 partition. You might wonder, <em is there any way I can see the state of the cluster and the partitions ? You can use the sinfo command
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ sinfo -M gpu
CLUSTER: gpu
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
gtx1080* up infinite 1 drain gpu-stage08
gtx1080* up infinite 13 mix gpu-n[16-19,22-25],gpu-stage[09-11,13-14]
gtx1080* up infinite 3 idle gpu-n[20-21],gpu-stage12
titanx up infinite 4 mix gpu-stage[02,04-06]
titanx up infinite 3 idle gpu-stage[01,03,07]
k40 up infinite 1 idle smpgpu-n0
v100 up infinite 1 mix gpu-n27
power9 up infinite 4 idle ppc-n[1-4]
scavenger up infinite 1 drain gpu-stage08
scavenger up infinite 18 mix gpu-n[16-19,22-25,27],gpu-stage[02,04-06,09-11,13-14]
scavenger up infinite 7 idle gpu-n[20-21],gpu-stage[01,03,07,12],smpgpu-n0
a100 up infinite 1 mix gpu-n28
a100 up infinite 2 idle gpu-n[29-30]
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$
To see all the cluster info, pass a comma separate list of cluster names to the -M flag
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ sinfo -M smp,gpu,mpi,htc
CLUSTER: gpu
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
gtx1080* up infinite 1 drain gpu-stage08
gtx1080* up infinite 13 mix gpu-n[16-19,22-25],gpu-stage[09-11,13-14]
gtx1080* up infinite 3 idle gpu-n[20-21],gpu-stage12
titanx up infinite 4 mix gpu-stage[02,04-06]
titanx up infinite 3 idle gpu-stage[01,03,07]
k40 up infinite 1 idle smpgpu-n0
v100 up infinite 1 mix gpu-n27
power9 up infinite 4 idle ppc-n[1-4]
scavenger up infinite 1 drain gpu-stage08
scavenger up infinite 18 mix gpu-n[16-19,22-25,27],gpu-stage[02,04-06,09-11,13-14]
scavenger up infinite 7 idle gpu-n[20-21],gpu-stage[01,03,07,12],smpgpu-n0
a100 up infinite 1 mix gpu-n28
a100 up infinite 2 idle gpu-n[29-30]
CLUSTER: htc
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
htc* up infinite 11 mix htc-n[28-29,100-103,107-110,112]
htc* up infinite 2 alloc htc-n[27,105]
htc* up infinite 29 idle htc-n[0-26,30-31]
scavenger up infinite 11 mix htc-n[28-29,100-103,107-110,112]
scavenger up infinite 2 alloc htc-n[27,105]
scavenger up infinite 29 idle htc-n[0-26,30-31]
CLUSTER: mpi
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
opa* up infinite 2 down* opa-n[63,77]
opa* up infinite 81 alloc opa-n[0-45,50-53,55-56,61-62,64-76,78-83,88-95]
opa* up infinite 12 idle opa-n[46-49,57-60,84-87]
opa* up infinite 1 down opa-n54
opa-high-mem up infinite 36 alloc opa-n[96-131]
ib up infinite 6 resv ib-n[0-3,12-13]
ib up infinite 12 alloc ib-n[4-5,7-11,18-19,26-28]
ib up infinite 14 idle ib-n[6,14-17,20-25,29-31]
scavenger up infinite 2 down* opa-n[63,77]
scavenger up infinite 117 alloc opa-n[0-45,50-53,55-56,61-62,64-76,78-83,88-131]
scavenger up infinite 12 idle opa-n[46-49,57-60,84-87]
scavenger up infinite 1 down opa-n54
CLUSTER: smp
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
smp* up infinite 3 down* smp-n[0,8,151]
smp* up infinite 124 mix smp-n[1,24-32,34-37,39-40,42-44,47-49,51-53,55,57-58,60,62-63,65-66,68-69,73-75,77,80-82,84-92,96-98,101-103,105-107,109-111,113-114,116,119,126-127,131-132,134-138,140,143-144,150,152-153,157-165,167-168,171,173-181,183-184,187,189-200,202,204-205,207-208,210]
smp* up infinite 49 alloc smp-n[2,4-6,11,13-14,16,20-21,23,33,38,41,50,54,56,59,61,64,67,70-71,78-79,99-100,104,108,112,115,121-122,129,133,139,142,145,154-156,166,169-170,182,185,188,201,206]
smp* up infinite 30 idle smp-n[3,7,9-10,12,15,19,22,45-46,72,76,83,93-95,117-118,120,128,130,141,146-149,172,186,203,209]
high-mem up infinite 6 mix smp-256-n[1-2],smp-3072-n[0-3]
high-mem up infinite 1 alloc smp-nvme-n1
high-mem up infinite 3 idle smp-512-n[1-2],smp-1024-n0
legacy up infinite 2 mix legacy-n[13,16]
legacy up infinite 5 alloc legacy-n[7-11]
legacy up infinite 12 idle legacy-n[0-6,14-15,17-19]
legacy up infinite 1 down legacy-n12
scavenger up infinite 3 down* smp-n[0,8,151]
scavenger up infinite 132 mix legacy-n[13,16],smp-256-n[1-2],smp-3072-n[0-3],smp-n[1,24-32,34-37,39-40,42-44,47-49,51-53,55,57-58,60,62-63,65-66,68-69,73-75,77,80-82,84-92,96-98,101-103,105-107,109-111,113-114,116,119,126-127,131-132,134-138,140,143-144,150,152-153,157-165,167-168,171,173-181,183-184,187,189-200,202,204-205,207-208,210]
scavenger up infinite 55 alloc legacy-n[7-11],smp-n[2,4-6,11,13-14,16,20-21,23,33,38,41,50,54,56,59,61,64,67,70-71,78-79,99-100,104,108,112,115,121-122,129,133,139,142,145,154-156,166,169-170,182,185,188,201,206],smp-nvme-n1
scavenger up infinite 45 idle legacy-n[0-6,14-15,17-19],smp-512-n[1-2],smp-1024-n0,smp-n[3,7,9-10,12,15,19,22,45-46,72,76,83,93-95,117-118,120,128,130,141,146-149,172,186,203,209]
scavenger up infinite 1 down legacy-n12
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$
You can use a similar syntax for the squeue command to see all the jobs you have submitted.
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ squeue -M smp,gpu,mpi,htc -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260052 titanx gpus-1 gnowmik R 14:46 1 gpu-stage06
CLUSTER: htc
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: mpi
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ sbatch amber.slurm
Submitted batch job 260055 on cluster gpu
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ cd ../mocvnhlysm_1N.24C_OMPI_SMP/
[gnowmik@login1 mocvnhlysm_1N.24C_OMPI_SMP]$ sbatch amber.slurm
[gnowmik@login1 mocvnhlysm_2GTX1080.2C]$ squeue -M smp,gpu,mpi,htc -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260055 gtx1080 gpus-2 gnowmik R 0:03 1 gpu-n25
260052 titanx gpus-1 gnowmik R 15:46 1 gpu-stage06
CLUSTER: htc
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: mpi
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
5105649 high-mem mocv gnowmik R 0:28 1 smp-512-n1
Next, I'm going to change the job submission script to submit to the v100 partition on the gpu cluster
[gnowmik@login1 ~]$ cp -rp mocvnhlysm_1titanX.1C mocvnhlysm_1v100.1C
[gnowmik@login1 ~]$ cd mocvnhlysm_1v100.1C
[gnowmik@login1 mocvnhlysm_1v100.1C]$ vi amber.slurm
[gnowmik@login1 mocvnhlysm_1v100.1C]$ head amber.slurm
#!/bin/bash
#SBATCH --job-name=gpus-1
#SBATCH --output=gpus-1.out
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cluster=gpu
#SBATCH --partition=v100
#SBATCH --gres=gpu:1
#SBATCH --time=24:00:00
[gnowmik@login1 mocvnhlysm_1v100.1C]$
[gnowmik@login1 mocvnhlysm_1v100.1C]$ sbatch amber.slurm
Submitted batch job 260056 on cluster gpu
[gnowmik@login1 mocvnhlysm_1v100.1C]$ squeue -M smp,gpu,mpi,htc -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260056 v100 gpus-1 gnowmik PD 0:00 1 (Priority)
260052 titanx gpus-1 gnowmik R 20:44 1 gpu-stage06
CLUSTER: htc
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: mpi
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
[gnowmik@login1 mocvnhlysm_1v100.1C]$
To obtain more information about why the job is in the PD state, use the scontrol command
[gnowmik@login1 mocvnhlysm_1v100.1C]$ scontrol -M gpu show job 260056
JobId=260056 JobName=gpus-1
UserId=gnowmik(152289) GroupId=kwong(16260) MCS_label=N/A
Priority=2367 Nice=0 Account=sam QOS=gpu-v100-s
JobState=PENDING Reason=Priority Dependency=(null)
Requeue=1 Restarts=0 BatchFlag=1 Reboot=0 ExitCode=0:0
RunTime=00:00:00 TimeLimit=1-00:00:00 TimeMin=N/A
SubmitTime=2022-01-26T08:20:43 EligibleTime=2022-01-26T08:20:43
AccrueTime=2022-01-26T08:20:43
StartTime=Unknown EndTime=Unknown Deadline=N/A
SuspendTime=None SecsPreSuspend=0 LastSchedEval=2022-01-26T08:24:34
Partition=v100 AllocNode:Sid=login1:25474
ReqNodeList=(null) ExcNodeList=(null)
NodeList=(null)
NumNodes=1-1 NumCPUs=1 NumTasks=1 CPUs/Task=1 ReqB:S:C:T=0:0:*:*
TRES=cpu=1,mem=5364M,node=1,billing=5,gres/gpu=1
Socks/Node=* NtasksPerN:B:S:C=1:0:*:* CoreSpec=*
MinCPUsNode=1 MinMemoryCPU=5364M MinTmpDiskNode=0
Features=(null) DelayBoot=00:00:00
OverSubscribe=OK Contiguous=0 Licenses=(null) Network=(null)
Command=/ihome/kwong/gnowmik/mocvnhlysm_1v100.1C/amber.slurm
WorkDir=/ihome/kwong/gnowmik/mocvnhlysm_1v100.1C
StdErr=/ihome/kwong/gnowmik/mocvnhlysm_1v100.1C/gpus-1.out
StdIn=/dev/null
StdOut=/ihome/kwong/gnowmik/mocvnhlysm_1v100.1C/gpus-1.out
Power=
TresPerNode=gpu:1
MailUser=(null) MailType=NONE
[gnowmik@login1 mocvnhlysm_1v100.1C]$
If you realize that you made a mistake in the inputs for your job submission script, you can cancel the job with the scancel command
[gnowmik@login1 mocvnhlysm_1v100.1C]$ squeue -M smp,gpu,mpi,htc -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260056 v100 gpus-1 gnowmik PD 0:00 1 (Priority)
260052 titanx gpus-1 gnowmik R 26:07 1 gpu-stage06
CLUSTER: htc
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: mpi
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
[gnowmik@login1 mocvnhlysm_1v100.1C]$ scancel -M gpu 260056
[gnowmik@login1 mocvnhlysm_1v100.1C]$ squeue -M smp,gpu,mpi,htc -u $USER
CLUSTER: gpu
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
260052 titanx gpus-1 gnowmik R 26:24 1 gpu-stage06
CLUSTER: htc
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: mpi
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
CLUSTER: smp
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
[gnowmik@login1 mocvnhlysm_1v100.1C]$
That's it! Once you become familiar with these handful of commands, you should become proficient in leveraging all the compute resources for your research. The hardest part is crafting the job submission script; however, CRC is building a collection of examples within the directory /ihome/crc/how_to_run/ that might address your specific application.
CRC provides a few helper scripts that are intended to make the user experience simpler. These include
- Checking on disk quota
[gnowmik@login1 ~]$ crc-quota.py
User: 'gnowmik'
-> ihome: 70.11 GB / 75.0 GB
Group: 'kwong'
-> bgfs: 35.91 GB / 5.0 TB
[gnowmik@login1 ~]$
- Checking on allocation utilization
[gnowmik@login1 ~]$ crc-usage.pl
|----------------------------------------------------------------------------------|
| Proposal End Date | 01/26/23 |
|----------------------------------------------------------------------------------|
| Cluster: smp, Available SUs: 50000 |
|--------------------|------------------------------|------------------------------|
| User | SUs Used | Percentage of Total |
|--------------------|------------------------------|------------------------------|
| dap195 | 0 | 0.00 |
| gnowmik | 0 | 0.00 |
| haggis97 | 0 | 0.00 |
|--------------------|------------------------------|------------------------------|
| Overall | 0 | 0.00 |
|--------------------|------------------------------|------------------------------|
|----------------------------------------------------------------------------------|
| Cluster: mpi, Available SUs: 0 |
|--------------------|------------------------------|------------------------------|
| User | SUs Used | Percentage of Total |
|--------------------|------------------------------|------------------------------|
| dap195 | 0 | N/A |
| gnowmik | 0 | N/A |
| haggis97 | 0 | N/A |
|--------------------|------------------------------|------------------------------|
| Overall | 0 | N/A |
|--------------------|------------------------------|------------------------------|
|----------------------------------------------------------------------------------|
| Cluster: gpu, Available SUs: 0 |
|--------------------|------------------------------|------------------------------|
| User | SUs Used | Percentage of Total |
|--------------------|------------------------------|------------------------------|
| dap195 | 0 | N/A |
| gnowmik | 0 | N/A |
| haggis97 | 0 | N/A |
|--------------------|------------------------------|------------------------------|
| Overall | 0 | N/A |
|--------------------|------------------------------|------------------------------|
|----------------------------------------------------------------------------------|
| Cluster: htc, Available SUs: 0 |
|--------------------|------------------------------|------------------------------|
| User | SUs Used | Percentage of Total |
|--------------------|------------------------------|------------------------------|
| dap195 | 0 | N/A |
| gnowmik | 0 | N/A |
| haggis97 | 0 | N/A |
|--------------------|------------------------------|------------------------------|
| Overall | 0 | N/A |
|--------------------|------------------------------|------------------------------|
| Aggregate |
|----------------------------------------|-----------------------------------------|
| Investments Total | 150000^a |
| Aggregate Usage (no investments) | 0.00 |
| Aggregate Usage | 0.00 |
|----------------------------------------|-----------------------------------------|
| ^a Investment SUs can be used across any cluster |
|----------------------------------------------------------------------------------|
[gnowmik@login1 ~]$
- Scanning for
idleresources
[gnowmik@login1 ~]$ crc-idle.py
Cluster: smp, Partition: smp
============================
2 nodes w/ 1 idle cores
5 nodes w/ 2 idle cores
1 nodes w/ 3 idle cores
9 nodes w/ 4 idle cores
2 nodes w/ 5 idle cores
11 nodes w/ 6 idle cores
30 nodes w/ 7 idle cores
35 nodes w/ 8 idle cores
1 nodes w/ 9 idle cores
11 nodes w/ 12 idle cores
4 nodes w/ 15 idle cores
1 nodes w/ 16 idle cores
1 nodes w/ 18 idle cores
1 nodes w/ 21 idle cores
1 nodes w/ 22 idle cores
20 nodes w/ 23 idle cores
Cluster: smp, Partition: high-mem
=================================
6 nodes w/ 8 idle cores
2 nodes w/ 12 idle cores
Cluster: smp, Partition: legacy
===============================
1 nodes w/ 1 idle cores
1 nodes w/ 8 idle cores
Cluster: gpu, Partition: gtx1080
================================
3 nodes w/ 1 idle GPUs
1 nodes w/ 2 idle GPUs
4 nodes w/ 3 idle GPUs
4 nodes w/ 4 idle GPUs
Cluster: gpu, Partition: titanx
===============================
1 nodes w/ 1 idle GPUs
1 nodes w/ 2 idle GPUs
1 nodes w/ 3 idle GPUs
3 nodes w/ 4 idle GPUs
Cluster: gpu, Partition: k40
============================
1 nodes w/ 2 idle GPUs
Cluster: gpu, Partition: v100
=============================
No idle GPUs
Cluster: mpi, Partition: opa
============================
No idle cores
Cluster: mpi, Partition: opa-high-mem
=====================================
No idle cores
Cluster: mpi, Partition: ib
===========================
14 nodes w/ 20 idle cores
Cluster: htc, Partition: htc
============================
2 nodes w/ 2 idle cores
1 nodes w/ 5 idle cores
1 nodes w/ 6 idle cores
1 nodes w/ 10 idle cores
3 nodes w/ 11 idle cores
1 nodes w/ 12 idle cores
20 nodes w/ 16 idle cores
4 nodes w/ 24 idle cores
1 nodes w/ 25 idle cores
1 nodes w/ 37 idle cores
5 nodes w/ 48 idle cores
[gnowmik@login1 ~]$
- Requesting an interactive session on a compute node
[gnowmik@login1 ~]$ crc-interactive.py --help
crc-interactive.py -- An interactive Slurm helper
Usage:
crc-interactive.py (-s | -g | -m | -i | -d) [-hvzo] [-t <time>] [-n <num-nodes>]
[-p <partition>] [-c <num-cores>] [-u <num-gpus>] [-r <res-name>]
[-b <memory>] [-a <account>] [-l <license>] [-f <feature>]
Positional Arguments:
-s --smp Interactive job on smp cluster
-g --gpu Interactive job on gpu cluster
-m --mpi Interactive job on mpi cluster
-i --invest Interactive job on invest cluster
-d --htc Interactive job on htc cluster
Options:
-h --help Print this screen and exit
-v --version Print the version of crc-interactive.py
-t --time <time> Run time in hours, 1 <= time <= 12 [default: 1]
-n --num-nodes <num-nodes> Number of nodes [default: 1]
-p --partition <partition> Specify non-default partition
-c --num-cores <num-cores> Number of cores per node [default: 1]
-u --num-gpus <num-gpus> Used with -g only, number of GPUs [default: 0]
-r --reservation <res-name> Specify a reservation name
-b --mem <memory> Memory in GB
-a --account <account> Specify a non-default account
-l --license <license> Specify a license
-f --feature <feature> Specify a feature, e.g. `ti` for GPUs
-z --print-command Simply print the command to be run
-o --openmp Run using OpenMP style submission
[gnowmik@login1 ~]$ crc-interactive.py -g -p titanx -n 1 -c 1 -u 1 -t 12
srun: job 260065 queued and waiting for resources
srun: job 260065 has been allocated resources
[gnowmik@gpu-stage06 ~]$ nvidia-smi
Wed Jan 26 08:42:04 2022
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 460.32.03 Driver Version: 460.32.03 CUDA Version: 11.2 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 GeForce GTX TIT... On | 00000000:02:00.0 Off | N/A |
| 48% 82C P2 236W / 250W | 794MiB / 12212MiB | 99% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 1 GeForce GTX TIT... On | 00000000:03:00.0 Off | N/A |
| 22% 28C P8 16W / 250W | 1MiB / 12212MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 2 GeForce GTX TIT... On | 00000000:81:00.0 Off | N/A |
| 22% 28C P8 15W / 250W | 1MiB / 12212MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 3 GeForce GTX TIT... On | 00000000:82:00.0 Off | N/A |
| 22% 27C P8 14W / 250W | 1MiB / 12212MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 23796 C pmemd.cuda 790MiB |
+-----------------------------------------------------------------------------+
[gnowmik@gpu-stage06 ~]$ exit
exit
[gnowmik@login1 ~]$
There are a few other helper scripts that you can see by typing crc- followed two strokes of the tab key
[gnowmik@login1 ~]$ crc-
crc-idle.py crc-job-stats.py crc-quota.py crc-scontrol.py crc-squeue.py crc-usage.pl
crc-interactive.py crc-proposal-end.py crc-scancel.py crc-sinfo.py crc-sus.py
[gnowmik@login1 ~]$ crc-
The best way to get help is to submit a help ticket. You should log in to the CRC website using your Pitt credentials first.